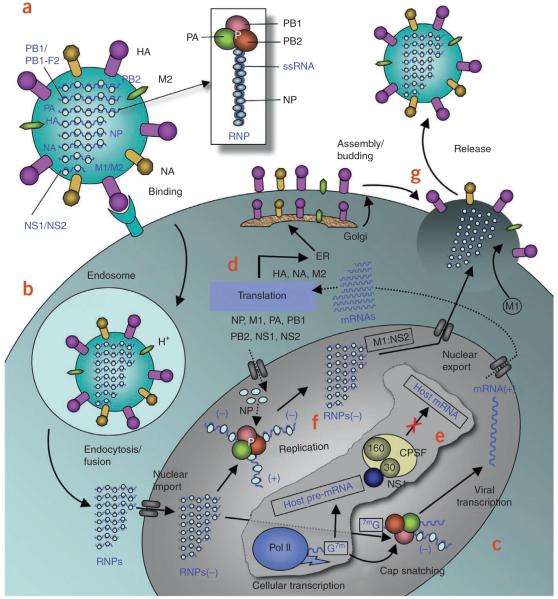
Figure 1.
Influenza A life cycle. (a) Influenza A virus has a lipid bilayer envelope, within which are eight RNA genomic segments, each of which is associated with the trimeric viral RNA polymerase (PB1, PB2, PA) and coated with multiple nucleoproteins (NPs) to form the vRNPs. The outer layer of the lipid envelope is spiked with multiple copies of HA, NA and a small number of M2, whereas the M1 molecules keep vRNPs attached to the inner layer. (b) The viral surface glycoprotein HA binds to the host cell-surface sialic acid receptors, and the virus is transported into the cell in an endocytic vesicle. The low pH in the endosome triggers a conformational change in the HA protein that leads to fusion of the viral and endosomal membranes. The low pH also triggers the flow of protons into the virus via the M2 ion channel, thereby dissociating the vRNPs from M1 matrix proteins. The vRNPs that are released into the cytoplasm are transported into the nucleus by recognition of the nuclear localization sequences (NLSs) on nucleoproteins83 only when the M1 molecules are dissociated. (c) In the nucleus, the viral polymerase initiates viral mRNA synthesis with 5′-capped RNA fragments cleaved from host pre-mRNAs. The PB2 subunit binds the 5′ cap of host pre-mRNAs84, and the endonuclease domain in PA subunit cleaves the pre-mRNA 10–13 nucleotides downstream from the cap85. Viral mRNA transcription is subsequently initiated from the cleaved 3′ end of the capped RNA segment85,86. This ‘cap snatching’ occurs on nascent pre-mRNAs. (d) Viral mRNAs are transported to the cytoplasm for translation into viral proteins. The surface proteins HA, M2 and NA are processed in the endoplasmic reticulum (ER), glycosylated in the Golgi apparatus and transported to the cell membrane. (e) The NS1 protein of influenza A virus serves a critical role in suppressing the production of host mRNAs by inhibiting the 3′-end processing of host pre-mRNAs60,87, consequently blocking the production of host mRNAs, including interferon-β mRNAs. Unlike host pre-mRNAs, the viral mRNAs do not require 3′-end processing by the host cell machinery. Therefore, the viral mRNAs are transported to the cytoplasm, whereas the host mRNA synthesis is predominantly blocked. (f) The viral polymerase is responsible for not only capped RNA-primed mRNA synthesis but also unprimed replication of vRNAs in steps (−) vRNA → (+) cRNA → (−) vRNA. The nucleoprotein molecules are required for these two steps of replication and are deposited on the cRNA and vRNA during RNA synthesis30. The resulting vRNPs are subsequently transported to the cytoplasm, mediated by a M1–NS2 complex that is bound to the vRNPs; NS2 interacts with human CRM1 protein that exports the vRNPs from the nucleus88. (g) The vRNPs reach the cell membrane to be incorporated into new viruses (reviewed in ref. 89) that are budded out. The HA and NA proteins in new viruses contain terminal sialic acids that would cause the viruses to clump together and adhere to the cell surface. The NA of newly formed viruses cleaves these sialic acid residues, thereby releasing the virus from the host cell.