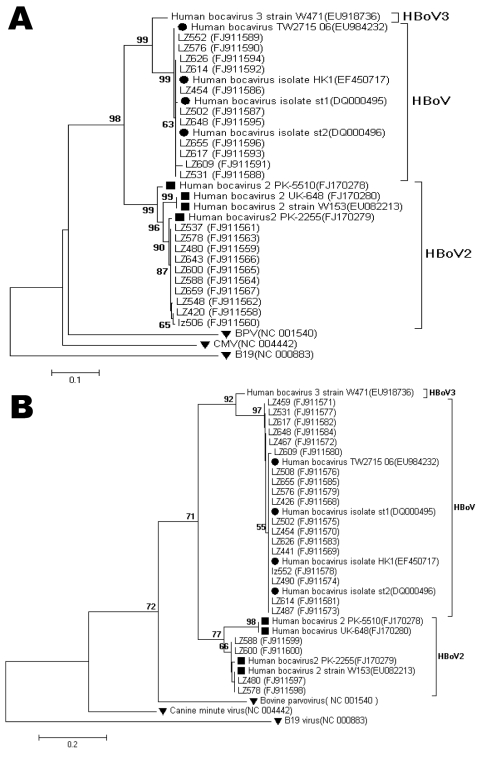
Figure.
Phylogenetic analysis of A) the partial nonstructural protein 1 (NS1) nucleotide sequences (412 bp) and B) the partial nucleoprotein 1 (NP1) nucleotide sequences (256 bp) of human bocavirus 2 (HBoV2), Gansu Province, People’s Republic of China. The phylogenetic trees were constructed by the neighbor-joining method using MEGA 3.1 (www.megasoftware.net), and bootstrap values were determined for 1,000 replicates. Bootstrap values >50% are shown at the branching points. Human bocavirus (HBoV) and HBoV2 reference sequences are indicated by circles and squares, respectively. Bovine parvovirus (BPV), cytomegalovirus (CMV), and human parvovirus B19 (B19) reference sequences are indicated by inverted triangles. Scale bars indicate nucleotide substitutions per site. Reference sequences were obtained from GenBank (accession nos. DQ000495, DQ000496, EF450717, EU082213, EU918736, EU984232, FJ170278, FJ170279, FJ170280, NC001540, NC004442, and NC000883). Sequences generated in this study were deposited in GenBank under accession nos. FJ911558–FJ911600.