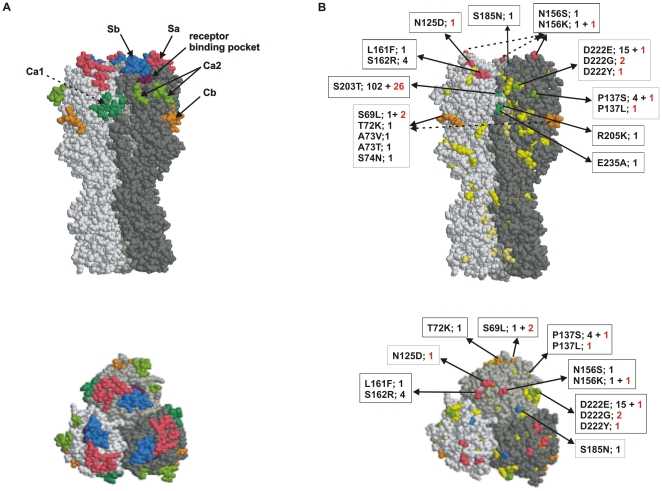
Figure 3. Amino acid differences in the HA between the Finnish pandemic viruses and the vaccine strain A/California/07/2009.
A. The trimeric HA molecule with previously identified H1 protein-related antigenic sites (Sa in pink, Sb in blue, Ca1 in darker green, Ca2 in lighter green and Cb in orange) of influenza A(H1N1) viruses and with the receptor binding pocket (purple) is presented in side and from top view. Different monomers are shown in various shades of grey color. The structure is based on the 3-dimensional structure of A/South Carolina/1/18 (RCSB Protein bank accession number 1ruz) HA molecule, which is genetically the closest resolved H1 structure as compared to the 2009 pandemic H1 molecule. B. The amino acid differences between the HA molecules of the Finnish pandemic viruses and the A/California/07/2009 vaccine virus are shown in the trimeric HA structure. Amino acid changes in antigenic sites are colored as in panel A. Other changes (apart from antigenic sites) in the HA1 region are shown in yellow and in the HA2 region in gold. Changes in antigenic sites are illustrated by the amino acid residue number, the amino acids that have changed, and in addition, the number of viruses found to contain the respective amino acid (numbers in red denote severe infections).