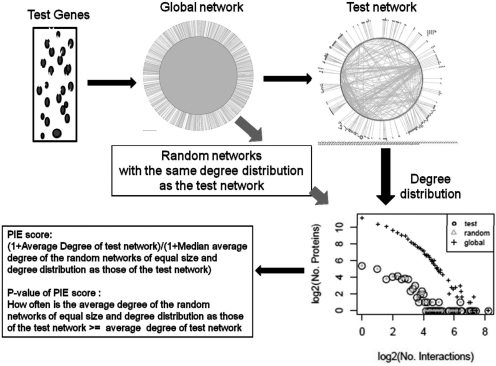
Fig. 2.
Summary of the PIE procedure. In the PIE procedure, a test set of genes of interest, e.g. those upregulated by a virus, are used to extract a specific PPI network from the general PPI network that consists of proteins encoded by genes in that test set. The number of interactions of each protein of the test set in the general PPI network (i.e. degree) is calculated. A number of proteins equal to the test set and with the same degree distribution as the test proteins in the general PPI network are then randomly selected from the set of all proteins in the general PPI network. As such the test set and the randomly selected set have the same degree distribution in the general PPI network. The average degree within the test set itself (i.e. total number of edges divided by total number of nodes in the test set) is then obtained and compared with that within the randomly selected set. The fraction of cases in which the average degree within a randomly selected set surpasses or equalizes that of the test network is recorded as the P-value for the physical cohesiveness of proteins of the genes of interest. The PIE score is the ratio of the average degree within the test network to the median average degree within the random networks.