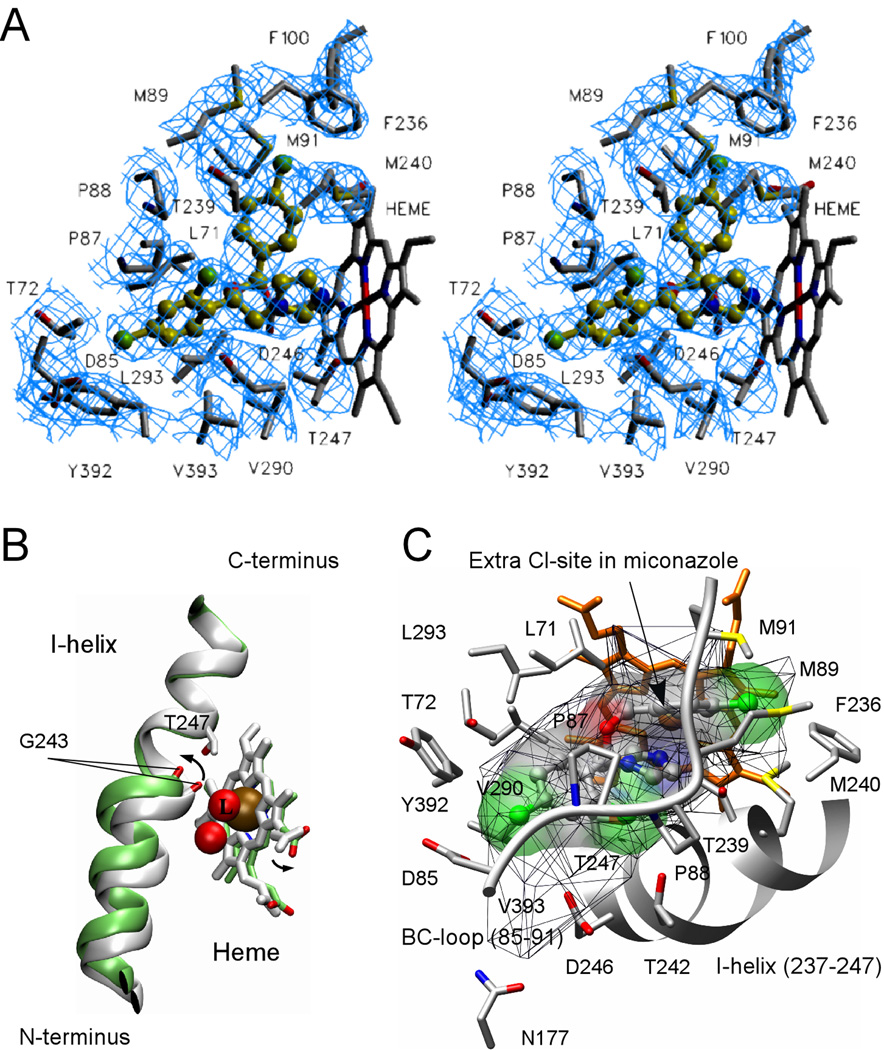
Fig. 7. Econazole binding in the active site.
A, stereo view of econazole (yellow-green) bound in the active site of CYP130 is shown. Cl atoms are colored in green, N atoms in blue and O atoms in red. Amino acid residues within 4 Å of econazole are labeled in black. Fragments of the 2Fo−Fc electron density composite omit map contoured at 1.0 σ are in blue. To avoid excessive cluttering, heme was excluded from the map calculation and T242 was excluded from the view as projecting on top of econazole. The image was generated using the SETOR program (52). B, a kink of the I-helix introduced by the binding of econazole is shown. The ligand-free (gray) and econazole-bound (green) CYP130 structures were superimposed with an r.m.s.d. of 0.93 Å2 for all the protein residues. Iron (ochre) and oxygen (red) atoms are shown as spheres. The iron axial water ligand is labeled with a capital L. The arrows show the directions of movements upon transition from the ligand-free to the econazole-bound state. C, alignment of the BC-loop fragment (85–91) (grey) in the groove formed between the mono- and double-chlorinated phenyl rings of econazole is shown. The additional chlorination site in miconazole is indicated by an arrow. The solid surface represents the van der Waals surface of econazole (volume of 330 Å3) and is colored according to the underlying atoms: oxygen in red, nitrogen in blue, chlorine in green. The mesh surface shows the accessible space in the active site (600 Å3). A fragment of the I-helix (237–247) is represented by the grey ribbon. The heme is in orange. This image was generated using the program CHIMERA (53).