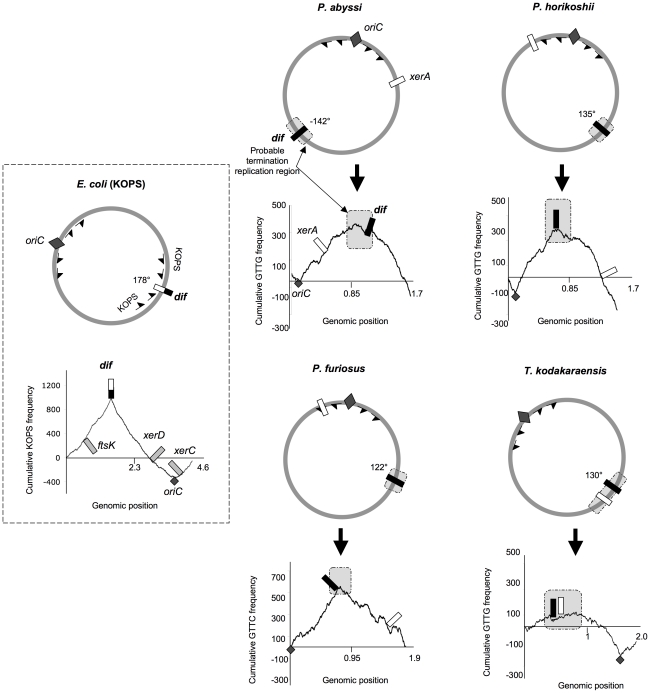
Figure 2. Genomic localization of dif sites and xer genes.
The E. coli KOPS cumulative skew produces a pyramidal shaped graphic with the dif site (black and white rectangle) located at the summit (178° from oriC). xerC, xerD and ftsK genes are located in the oriC region. ASPS skew graphics from Thermococcales show rounded shapes where oriC (dark gray diamond) can be localized precisely and where putative regions of replication termination (light gray rectangles) can be broadly determined. GTTG is the most skewed 4 nt sequence in P. abyssi, P. horikoshii and T. kodakaraensis genomes whereas GTTC is the most skewed 4 nt sequence in P. furiosus genome. Thermococcales dif sites (black rectangles) are all located at the summit of the ASPS skews graphics, at 120–142° from oriC, in putative replication termination regions. The xerA gene (white rectangles) positions are more variable. Genomic coordinates of oriC, dif and xerA genes can be found in Table S1.