Abstract
The dehydratase (DH) domain of module 4 of the 6-deoxyerythronolide B synthase (DEBS) has been shown to catalyze an exclusive syn elimination/syn addition of water. Incubation of recombinant DH4 with chemoenzymatically prepared anti-(2R,3R)-2-methyl-3-hydroxypentanoyl-ACP (2a-ACP) gave the dehydration product 3-ACP. Similarly, incubation of DH4 with synthetic 3-ACP resulted in the reverse enzyme-catalyzed hydration reaction, giving a ~3:1 equilbrium mixture of 2a-ACP and 3-ACP. Incubation of a mixture of propionyl-SNAC (4), methylmalonyl-CoA, and NADPH with the DEBS β-ketoacyl synthase – acyl transferase [KS6][AT6] didomain, DEBS ACP6, and the ketoreductase domain from tylactone synthase module 1 (TYLS KR1) generated in situ anti-2a-ACP that underwent DH4-catalyzed syn dehydration to give 3-ACP. DH4 did not dehydrate either syn-(2S,3R)-2b-ACP, syn-(2R,3S)-2c-ACP, or anti-(2S,3S)-2d-ACP generated in situ by DEBS KR1, DEBS KR6, or the rifamycin synthase KR7 (RIFS KR7), respectively. Similarly, incubation of a mixture of (2S,3R)-2-methyl-3-hydroxypentanoyl-N-acetylcysteamine thioester (2b-SNAC), methylmalonyl-CoA, and NADPH with DEBS [KS6][AT6], DEBS ACP6, and TYLS KR1 gave anti-(2R,3R)-6-ACP that underwent syn dehydration catalyzed by DEBS DH4 to give (4R,5R)-(E)-2,4-dimethyl-5-hydroxy-hept-2-enoyl-ACP (7-ACP). The structure and stereochemistry of 7 were established by GC-MS and LC-MS comparison of the derived methyl ester 7-Me to a synthetic sample of 7-Me.
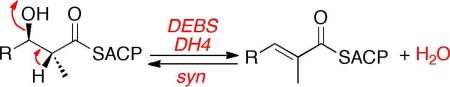
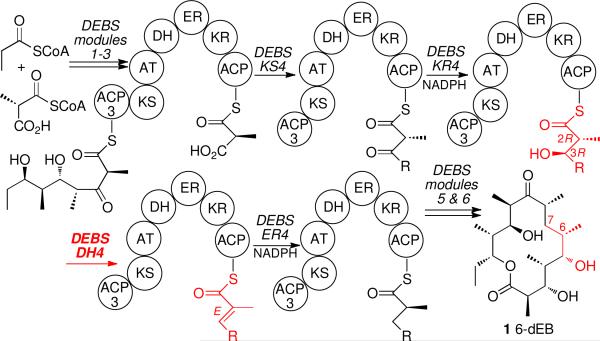
Of the more than 2000 non-aromatic polyketides, the vast majority contain one or more disubstituted or trisubstituted double bonds, most of which have E (trans) geometry.1 Moreover, essentially all polyketides that do not themselves display a double bond are biosynthesized by way of one or more unsaturated polyketide chain elongation intermediates. Thus although 6-deoxyerythronolide B (1, 6-dEB), the parent aglycone of the erythromycin family of antibiotics, does not have any double bonds in the final macrolactone, the responsible modular polyketide synthase (PKS), 6-dEB synthase (DEBS), does in fact harbor a dehydratase domain in module 4, termed DEBS DH4 (Figure 1).2,3,
Figure 1.
Proposed tetraketide substrate and pentaketide intermediates of DEBS module 4. The module has a KR, a DH and an ER domain in addition to the obligate KS, AT and ACP domains.
Direct evidence for the intermediacy of an unsaturated polyketide in erythromycin biosynthesis first came from disruption of the NADPH-binding motif of the ER4 domain, resulting in accumulation of a derivative of the corresponding (E)-Δ6,7-anhydro-6-dEB by mutants of the erythromycin producer Saccharopolyspora erythraea.4 Although the stereochemistry of the substrate for the DEBS DH4 dehydratase is not known, the responsible ketoreductase, DEBS KR4, is predicted to generate the (3R)-diastereomer of the 2-methyl-3-hydroxyacyl-ACP pentaketide, as deduced from the presence of a Leu-Ala-Asp triad closely correlated with the formation of (3R)-3-hydroxyacyl-ACP polyketide intermediates.5 Indeed, the vast majority of KR domains that are paired with a DH domain appear to harbor a conserved “Leu-Asp-Asp” motif.5a,b DEBS KR4 is also predicted to belong to the class of non-epimerizing ketoreductases, which would give rise to a (2R)-methyl group in the reduced product.5c
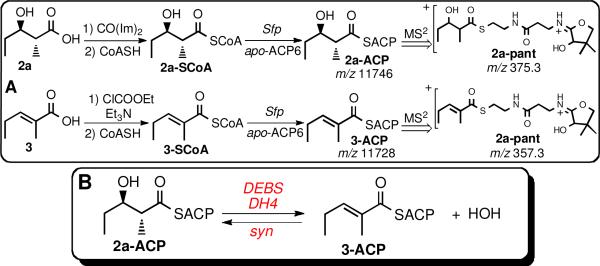
To establish the substrate specificity and stereochemical course of the DEBS DH4-catalyzed dehydration we used a chemoenzymatic strategy to prepare the requisite ACP-bound substrate and product analogues, 2a-ACP and 3-ACP. To this end the free acids 2a and 3 were each converted to the corresponding –SCoA thioesters, 2a-SCoA and 3-SCoA, and thence to anti-(2R,3R)-2-methyl-3-hydroxypentanoyl-ACP6 (2a-ACP) and the expected dehydration product, (E)-2-methylpent-2-enoyl-ACP (3-ACP), from DEBS apo-ACP6 using the phosphopantetheinyl transferase Sfp (Scheme 1A).6 The two ACP derivatives, which were readily distinguished by reverse phase LC-ESI(+)-MS, both exhibited the expected molecular weights.7 The structures were each confirmed by the MS2 phosphopantetheinate (PPant) ejection method which gave 2a-pant, m/z 375.33, and 3-pant, m/z 357.3, each with the predicted MW, as well as MS3 analysis of each of the characteristic PPant ejection fragments.8
Scheme 1.
A. Synthesis and analysis of ACP-bound substrates. B. DEBS DH4-catalyzed interconversion of 2a-ACP and 3-ACP.
Incubation of recombinant DEBS DH49 with 2a-ACP resulted in formation of the predicted dehydration product 3-ACP, as established by direct monitoring by LC-ESI(+)-MS3, including detection of the corresponding intact acyl-ACP and PPant ejection fragments for both 2a-ACP and 3-ACP (Scheme 1B). Similarly, incubation of DEBS DH4 with 3-ACP resulted in the reverse enzyme-catalyzed hydration reaction, giving a ~3:1 equilibrium mixture of 2a-ACP and 3-ACP.10
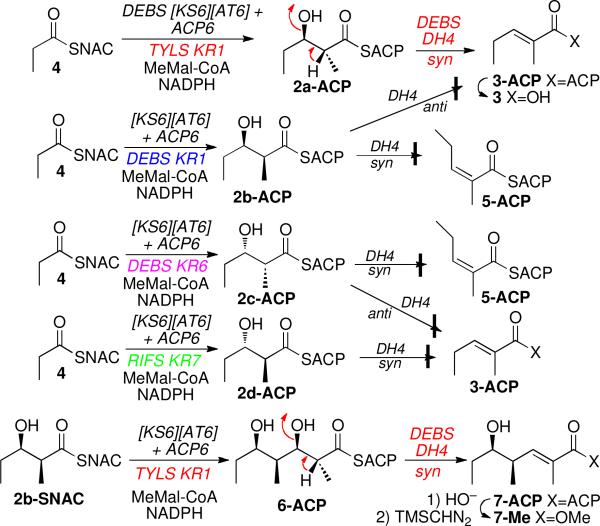
We also carried out combinatorial incubations using mixtures of recombinant PKS domains in order to generate in situ each of the 4 diastereomers of 2a–2d-ACP.11 In this manner, a mixture of the DEBS [KS6][AT6] didomain, DEBS ACP6, and TYLS KR1, the ketoreductase domain from module 1 of the tylactone synthase, was incubated with propionyl-SNAC (4), methylmalonyl-CoA, and NADPH to produce anti-(2R,3R)-2a-ACP.11b Addition of recombinant DEBS DH4, either simultaneous with or subsequent to the formation of 2a-ACP, resulted in dehydration to yield exclusively the predicted (E)-2-methylpent-2-enoyl-ACP (3-ACP), as confirmed by GC-MS analysis of the corresponding acid 3 and comparison with synthetic 3.12 By contrast, DEBS DH4 did not dehydrate either syn-(2S,3R)-2b-ACP or syn-(2R,3S)-2c-ACP generated by DEBS KR1 or KR6, respectively,11a,c to either E-3-ACP or the corresponding Z isomer 5-ACP, nor did DEBS DH4 dehydrate anti-(2S,3S)-2d-ACP produced by recombinant RIFS KR7,13 the KR domain from module 7 of the rifamycin synthase.
In further confirmation of the stereochemistry of the dehydration reaction, incubation of DEBS DH4 with anti-(2R,3R,4S,5R)-2,4-dimethyl-3,5-dihydroxyheptanoyl-ACP (6-ACP), generated in situ from 2b-SNAC, methylmalonyl-CoA, and NADPH by DEBS [KS6][AT6] + ACP6 + TYLS KR1, as previously described,11b gave exclusively E-7-ACP. The structure and stereochemistry of 7-ACP were determined by chiral GC-MS and LC-MS analysis of the derived methyl ester 7-Me, obtained by basic hydrolysis and treatment of the liberated acid with TMS-diazomethane, and comparison with an authentic synthetic standard of 7-Me.14
Sequence alignments of the DEBS DH4 domain with numerous PKS and FAS DH domains reveal conserved 2409HXXXGXXXXP and 2571D(A/V)(V/A)(A/L)(Q/H) motifs.2 Site-directed mutagenesis of the conserved active site His2409 of the DEBS DH4 domain abolished DEBS activity in Sac. Erythraea15a while the analogous His mutation also inactivates the homologous DH2 domain of the picromycin synthase.15b Together the conserved His and Asp residues comprise the catalytic dyad of the dehydratase, in which the active site His acts as a general base while the Asp2571, located 4.1Å from H2409 at the base of the substrate tunnel, is thought to serve as a general acid.9,16,17,,
Our results establish definitively that the DEBS DH4 domain catalyzes a syn elimination of water during erythromycin biosynthesis. The prototype dehydration catalyzed by the DH domain of the yeast FAS to give the characteristic disubstituted (E)-enoyl-ACP intermediates of fatty acid biosynthesis also takes place with net syn stereochemistry,18 as do the dehydrations catalyzed by the DH domains of module 2 of nanchangmycin synthase19 and module 2 of tylactone synthase.11b Indeed, the significant levels of overall sequence identity (>40%) and similarity (>55%) and the presence of the conserved motifs containing the catalytic dyad in more than 50 DH domains from a wide range of modular PKS systems, strongly suggest that the formation of all (E)-unsaturated polyketide intermediates involves a common syn dehydration mechanism.
Supplementary Material
Scheme 2.
Stereochemistry of DEBS DH4-catalyzed dehydration.
Acknowledgment
This work was supported by NIH grants GM22172 (D.E.C.) and CA66736 (C.K.) and a Welch Foundation Grant (A.K.C).
Footnotes
Supporting Information Available: Experimental procedures, LC-ESI(+)-MS3, and GC-MS data This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Shiomi K, Omura S. In Macrolide Antibiotics. In: Omura S, editor. Chemistry, Biology, and Practice. Second ed. Academic Press; San Diego, CA: 2002. pp. 1–56. [Google Scholar]
- 2.a Donadio S, Staver MJ, Mcalpine JB, Swanson SJ, Katz L. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]; Donadio S, Staver MJ, Mcalpine JB, Swanson SJ, Katz L. Gene. 1992;115:97–103. doi: 10.1016/0378-1119(92)90546-2. [DOI] [PubMed] [Google Scholar]; b Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 3.Each PKS module typically has 3 core catalytic domains: the β-ketoacyl-ACP synthase (KS), the acyltransferase (AT), and the acyl carrier protein (ACP). Most modules also carry specific combinations of ketoreductase (KR), dehydratase (DH) and enoylreductase (ER) tailoring domains.
- 4.Donadio S, McAlpine JB, Sheldon PJ, Jackson M, Katz L. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7119–7123. doi: 10.1073/pnas.90.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Reid R, Piagentini M, Rodriguez E, Ashley G, Viswanathan N, Carney J, Santi DV, Hutchinson CR, McDaniel R. Biochemistry. 2003;42:72–79. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]; b Caffrey P. ChemBioChem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]; c Keatinge-Clay AT. Chem. Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]; Keatinge-Clay AT, Stroud RM. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 7.Although it is not essential to the application of the ppant ejection methodology, 2a-ACP and 3-ACP were cleanly resolved under the LC conditions used.
- 8.Dorrestein PC, Bumpus SB, Calderone CT, Garneau-Tsodikova S, Aron ZD, Straight PD, Kolter R, Walsh CT, Kelleher NL. Biochemistry. 2006;45:12756–12766. doi: 10.1021/bi061169d. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Meluzzi D, Zheng WH, Hensler M, Nizet V, Dorrestein PC. Bioorg. Med. Chem. Lett. 2008;18:3107–3111. doi: 10.1016/j.bmcl.2007.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keatinge-Clay A. J. Mol. Biol. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The equilibrium for the FAS catalyzed dehydration to a disubstituted enoyl-ACP strong also favors the hydrated form by ~3:1. Cf Brown A, Affleck V, Kroon J, Slabas A. FEBS Lett. 2009;583:363–368. doi: 10.1016/j.febslet.2008.12.022.
- 11.a Castonguay R, He W, Chen AY, Khosla C, Cane DE. J. Am. Chem. Soc. 2007;129:13758–13769. doi: 10.1021/ja0753290. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Castonguay R, Valenzano CR, Chen AY, Keatinge-Clay A, Khosla C, Cane DE. J. Am. Chem. Soc. 2008;130:11598–11599. doi: 10.1021/ja804453p. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Valenzano CR, Lawson RJ, Chen AY, Khosla C, Cane DE. J. Am. Chem. Soc. 2009;131:18501–18511. doi: 10.1021/ja908296m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The E-isomer 3 was cleanly resolved from the Z isomer 5 and the two compounds were not interconverted under the conditions of the reaction.
- 13.You Y-O, Cane DE. unpublished observations.
- 14.Although DH4 processed both 2a-ACP and 6-ACP, DEBS DH4 does not dehydrate the corresponding SNAC thioester analog, anti-(2R,3R)-2a-SNAC (ref 9). This observation is consistent with the presence of a presumptive ACP-binding region on the DH surface adjacent to the entrance to the substrate binding tunnel, a feature also found in bacterial FabZ proteins (ref 17).
- 15.a Bevitt DJ, Staunton J, Leadlay PF. Biochem. Soc. Trans. 1993;21:30S. doi: 10.1042/bst021030s. [DOI] [PubMed] [Google Scholar]; b Wu J, Zaleski TJ, Valenzano C, Khosla C, Cane DE. J. Am. Chem. Soc. 2005;127:17393–17404. doi: 10.1021/ja055672+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.In the corresponding Type II FAS FabZ domains, a conserved Glu serves the same role as this Asp residue (ref 17).
- 17.Kimber MS, Martin F, Lu Y, Houston S, Vedadi M, Dharamsi A, Fiebig KM, Schmid M, Rock CO. J. Biol. Chem. 2004;279:52593–52602. doi: 10.1074/jbc.M408105200. [DOI] [PubMed] [Google Scholar]; Swarnamukhi PL, Sharma SK, Bajaj P, Surolia N, Surolia A, Suguna K. FEBS letters. 2006;580:2653–2660. doi: 10.1016/j.febslet.2006.04.014. [DOI] [PubMed] [Google Scholar]; Zhang L, Liu W, Hu T, Du L, Luo C, Chen K, Shen X, Jiang H. J. Biol. Chem. 2008;283:5370–5379. doi: 10.1074/jbc.M705566200. [DOI] [PubMed] [Google Scholar]
- 18.Sedgwick B, Morris C, French SJJCS. Chem. Commun. 1978:193–194. [Google Scholar]
- 19.Guo X, Liu T, Valenzano C, Deng Z, Cane DE. J. Am. Chem. Soc. 2010 doi: 10.1021/ja1073432. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.