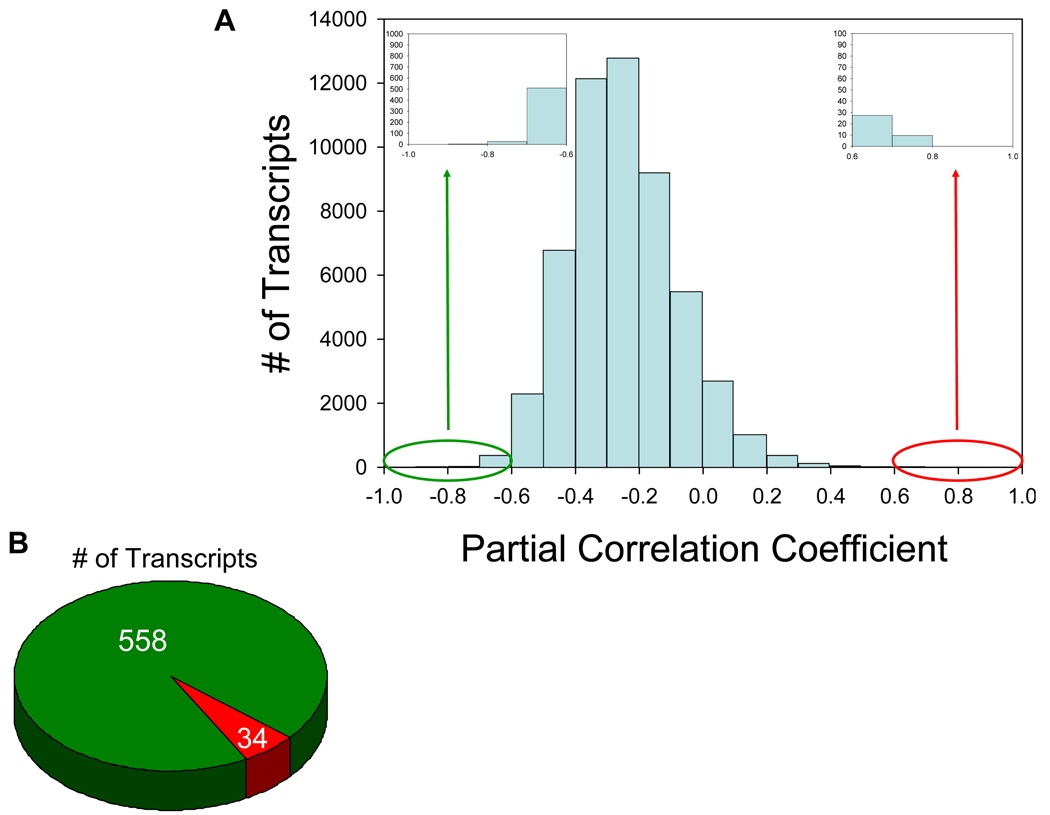
Figure 1. The majority of transcripts in lymphatic tissue are negatively associated with viral load.
(A) Histogram of the partial correlation coefficients for each transcript in the Affymetrix Human Genome U133 Plus 2.0 array. The partial correlation coefficient represents the strength of association between viral load and gene expression while controlling for stage of disease. Transcripts with an absolute value exceeding 0.6 were deemed differentially expressed while controlling for a false discovery rate of 0.7. (B) Pie chart depicting the number of transcripts remaining after applying the selection criteria (−0.6 > partial correlation coefficient > 0.6; 7% false discovery rate). The green and red sectors identify transcripts with a negative and positive association, respectively, between gene expression and viral load.