Abstract
In the crystal structure of the title compound, C8H10N2O3, molecules are linked by N—H⋯O hydrogen bonds, forming ribbons of centrosymmetric dimers extending along the c axis.
Related literature
For related literature, see: Banwell et al. (2006 ▶); Bernstein et al. (1995 ▶); Faulkner (2002 ▶); Sosa et al. (2002 ▶); Zeng (2006 ▶); Zeng et al. (2007 ▶).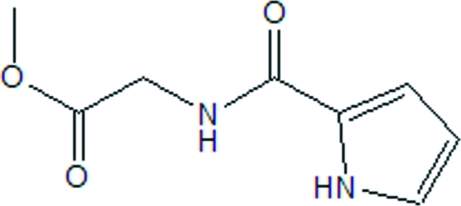
Experimental
Crystal data
C8H10N2O3
M r = 182.18
Monoclinic,

a = 11.3398 (19) Å
b = 5.0732 (9) Å
c = 16.500 (3) Å
β = 108.060 (3)°
V = 902.5 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 173 (2) K
0.48 × 0.41 × 0.21 mm
Data collection
Bruker SMART 1K CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1997 ▶) T min = 0.952, T max = 0.979
4219 measured reflections
1576 independent reflections
1417 reflections with I > 2σ(I)
R int = 0.033
Refinement
R[F 2 > 2σ(F 2)] = 0.052
wR(F 2) = 0.166
S = 1.10
1576 reflections
119 parameters
H-atom parameters constrained
Δρmax = 0.21 e Å−3
Δρmin = −0.31 e Å−3
Data collection: SMART (Bruker, 1999 ▶); cell refinement: SAINT-Plus (Bruker, 1999 ▶); data reduction: SAINT-Plus; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808027451/cf2214sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808027451/cf2214Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Enhanced figure: interactive version of Fig. 1
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1i | 0.88 | 1.93 | 2.782 (2) | 162 |
| N2—H2⋯O2ii | 0.88 | 2.09 | 2.9372 (19) | 161 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
We thank the Natural Science Foundation of Guangdong Province, China (No. 06300581) for generously supporting this study.
supplementary crystallographic information
Comment
Pyrrole derivatives are well known in many marine organisms (Faulkner, 2002). Some show important bioactivities, such as antitumor activity (Banwell et al., 2006) and protein kinase inhibiting activity (Sosa et al., 2002). This is the reason why they have attracted our interest. This study follows our previous studies on methyl 2-(4,5-dibromo-1H-pyrrole-2-carboxamido)propionate (Zeng et al., 2007) and 3-bromo-1-methyl-6,7-dihydropyrrolo[2,3-c]azepine- 4,8(1H,5H)-dione (Zeng, 2006).
In the crystal structure, molecules of the title compound are linked through N1—H1···O1i hydrogen bonds to form centrosymmetric dimers (Fig. 2) of graph-set motif R22(10) (Bernstein et al., 1995), which are linked by N2—H2···O2ii hydrogen bonds, generating ribbons extending along the c axis (also shown in Fig. 2). Bond lengths and angles are unexceptional.
Experimental
The hydrochloric acid salt of glycine methyl ester (0.63 g, 5 mmol) and 2-trichloroacetylpyrrole (1.06 g, 5 mmol) were added to acetonitrile (12 ml), followed by the dropwise addition of triethylamine (1.4 ml). The mixture was stirred at room temperature for 10 h and then poured into water. After filtration, the precipitate was collected as a yellow solid. The impure product was dissolved in EtOH at room temperature. Light-yellow monoclinic crystals suitable for X-ray analysis (m.p. 420 K, 95.6% yield) grew over a period of one week when the solution was exposed to the air. CH&N elemental analysis. Calc. for C8H10N2O3: C 52.74, H 5.53, N 15.38%; found: C 52.78, H 5.59, N 15.49%.
Refinement
H atoms were positioned geometrically [C—H = 0.99Å for CH2, 0.98Å for CH3, 0.95Å for CH (aromatic), and N—H = 0.88 Å] and refined using a riding model, with Uiso = 1.2Ueq (1.5Ueq for the methyl group) of the parent atom.
Figures
Fig. 1.
The molecular structure of the title compound, with the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
Ribbons of dimers formed by hydrogen bonds (dashed lines).
Crystal data
| C8H10N2O3 | Dx = 1.341 Mg m−3 |
| Mr = 182.18 | Melting point: 420 K |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.3398 (19) Å | Cell parameters from 3468 reflections |
| b = 5.0732 (9) Å | θ = 2.6–27.0° |
| c = 16.500 (3) Å | µ = 0.10 mm−1 |
| β = 108.060 (3)° | T = 173 K |
| V = 902.5 (3) Å3 | Block, light yellow |
| Z = 4 | 0.48 × 0.41 × 0.21 mm |
| F(000) = 384 |
Data collection
| Bruker SMART 1K CCD area-detector diffractometer | 1576 independent reflections |
| Radiation source: fine-focus sealed tube | 1417 reflections with I > 2σ(I) |
| graphite | Rint = 0.033 |
| φ and ω scans | θmax = 25.0°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1997) | h = −13→13 |
| Tmin = 0.952, Tmax = 0.979 | k = −6→5 |
| 4219 measured reflections | l = −16→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.052 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.166 | H-atom parameters constrained |
| S = 1.10 | w = 1/[σ2(Fo2) + (0.1092P)2 + 0.2414P] where P = (Fo2 + 2Fc2)/3 |
| 1576 reflections | (Δ/σ)max = 0.001 |
| 119 parameters | Δρmax = 0.21 e Å−3 |
| 0 restraints | Δρmin = −0.31 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.89632 (13) | 0.2572 (2) | 0.45357 (8) | 0.0387 (4) | |
| O2 | 0.63332 (13) | 0.2895 (3) | 0.31013 (8) | 0.0391 (4) | |
| N2 | 0.85781 (14) | 0.5697 (3) | 0.35256 (9) | 0.0343 (4) | |
| H2 | 0.8777 | 0.6472 | 0.3109 | 0.041* | |
| O3 | 0.58657 (13) | 0.4945 (3) | 0.41547 (9) | 0.0513 (5) | |
| C3 | 1.06971 (17) | 0.3350 (4) | 0.29921 (12) | 0.0364 (5) | |
| H3 | 1.0414 | 0.4778 | 0.2608 | 0.044* | |
| N1 | 1.08932 (14) | 0.0465 (3) | 0.40331 (10) | 0.0343 (4) | |
| H1 | 1.0773 | −0.0386 | 0.4466 | 0.041* | |
| C5 | 0.92253 (15) | 0.3607 (3) | 0.39334 (10) | 0.0306 (5) | |
| C6 | 0.75496 (17) | 0.6645 (3) | 0.37859 (12) | 0.0352 (5) | |
| H6A | 0.7211 | 0.8266 | 0.3464 | 0.042* | |
| H6B | 0.7847 | 0.7099 | 0.4400 | 0.042* | |
| C7 | 0.65395 (17) | 0.4608 (3) | 0.36324 (11) | 0.0329 (5) | |
| C4 | 1.02305 (17) | 0.2601 (3) | 0.36367 (11) | 0.0313 (5) | |
| C1 | 1.17634 (18) | −0.0142 (4) | 0.36582 (13) | 0.0394 (5) | |
| H1A | 1.2344 | −0.1546 | 0.3817 | 0.047* | |
| C2 | 1.16634 (18) | 0.1618 (4) | 0.30085 (13) | 0.0413 (5) | |
| H2A | 1.2159 | 0.1652 | 0.2638 | 0.050* | |
| C8 | 0.4845 (3) | 0.3088 (6) | 0.40341 (18) | 0.0720 (9) | |
| H8A | 0.4282 | 0.3231 | 0.3450 | 0.108* | |
| H8B | 0.4391 | 0.3494 | 0.4437 | 0.108* | |
| H8C | 0.5174 | 0.1290 | 0.4135 | 0.108* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0451 (8) | 0.0384 (8) | 0.0386 (8) | 0.0040 (6) | 0.0217 (6) | 0.0103 (5) |
| O2 | 0.0448 (8) | 0.0368 (7) | 0.0392 (8) | −0.0018 (6) | 0.0180 (6) | −0.0094 (6) |
| N2 | 0.0403 (9) | 0.0297 (8) | 0.0388 (9) | 0.0010 (6) | 0.0207 (7) | 0.0065 (6) |
| O3 | 0.0508 (9) | 0.0644 (10) | 0.0499 (9) | −0.0184 (7) | 0.0319 (8) | −0.0247 (7) |
| C3 | 0.0388 (10) | 0.0350 (10) | 0.0387 (10) | −0.0025 (8) | 0.0166 (8) | 0.0063 (8) |
| N1 | 0.0376 (9) | 0.0304 (8) | 0.0385 (9) | −0.0018 (6) | 0.0170 (7) | 0.0044 (6) |
| C5 | 0.0330 (9) | 0.0288 (10) | 0.0315 (9) | −0.0055 (7) | 0.0120 (8) | 0.0008 (7) |
| C6 | 0.0423 (11) | 0.0274 (9) | 0.0394 (10) | 0.0022 (7) | 0.0178 (8) | −0.0003 (7) |
| C7 | 0.0383 (10) | 0.0322 (9) | 0.0304 (9) | 0.0046 (7) | 0.0141 (8) | −0.0008 (7) |
| C4 | 0.0341 (9) | 0.0270 (9) | 0.0337 (10) | −0.0042 (7) | 0.0119 (8) | 0.0006 (7) |
| C1 | 0.0368 (10) | 0.0345 (10) | 0.0502 (12) | 0.0011 (8) | 0.0184 (9) | 0.0004 (8) |
| C2 | 0.0416 (11) | 0.0416 (11) | 0.0492 (12) | −0.0029 (8) | 0.0265 (9) | 0.0022 (9) |
| C8 | 0.0677 (16) | 0.097 (2) | 0.0690 (16) | −0.0400 (15) | 0.0468 (14) | −0.0364 (15) |
Geometric parameters (Å, °)
| O1—C5 | 1.239 (2) | N1—H1 | 0.880 |
| O2—C7 | 1.204 (2) | C5—C4 | 1.465 (2) |
| N2—C5 | 1.345 (2) | C6—C7 | 1.505 (3) |
| N2—C6 | 1.444 (2) | C6—H6A | 0.990 |
| N2—H2 | 0.880 | C6—H6B | 0.990 |
| O3—C7 | 1.328 (2) | C1—C2 | 1.373 (3) |
| O3—C8 | 1.458 (3) | C1—H1A | 0.950 |
| C3—C4 | 1.380 (2) | C2—H2A | 0.950 |
| C3—C2 | 1.398 (3) | C8—H8A | 0.980 |
| C3—H3 | 0.950 | C8—H8B | 0.980 |
| N1—C1 | 1.353 (2) | C8—H8C | 0.980 |
| N1—C4 | 1.365 (2) | ||
| C5—N2—C6 | 118.64 (14) | O2—C7—O3 | 122.96 (17) |
| C5—N2—H2 | 120.7 | O2—C7—C6 | 125.73 (17) |
| C6—N2—H2 | 120.7 | O3—C7—C6 | 111.30 (15) |
| C7—O3—C8 | 114.87 (16) | N1—C4—C3 | 107.59 (16) |
| C4—C3—C2 | 107.31 (17) | N1—C4—C5 | 119.11 (15) |
| C4—C3—H3 | 126.3 | C3—C4—C5 | 133.30 (17) |
| C2—C3—H3 | 126.3 | N1—C1—C2 | 108.28 (17) |
| C1—N1—C4 | 109.46 (15) | N1—C1—H1A | 125.9 |
| C1—N1—H1 | 125.3 | C2—C1—H1A | 125.9 |
| C4—N1—H1 | 125.3 | C1—C2—C3 | 107.37 (17) |
| O1—C5—N2 | 120.38 (16) | C1—C2—H2A | 126.3 |
| O1—C5—C4 | 121.72 (16) | C3—C2—H2A | 126.3 |
| N2—C5—C4 | 117.89 (14) | O3—C8—H8A | 109.5 |
| N2—C6—C7 | 111.34 (14) | O3—C8—H8B | 109.5 |
| N2—C6—H6A | 109.4 | H8A—C8—H8B | 109.5 |
| C7—C6—H6A | 109.4 | O3—C8—H8C | 109.5 |
| N2—C6—H6B | 109.4 | H8A—C8—H8C | 109.5 |
| C7—C6—H6B | 109.4 | H8B—C8—H8C | 109.5 |
| H6A—C6—H6B | 108.0 | ||
| C6—N2—C5—O1 | −2.1 (2) | C2—C3—C4—N1 | −0.2 (2) |
| C6—N2—C5—C4 | 177.39 (15) | C2—C3—C4—C5 | −179.42 (19) |
| C5—N2—C6—C7 | −63.6 (2) | O1—C5—C4—N1 | 0.2 (3) |
| C8—O3—C7—O2 | −0.4 (3) | N2—C5—C4—N1 | −179.30 (14) |
| C8—O3—C7—C6 | 178.56 (19) | O1—C5—C4—C3 | 179.36 (19) |
| N2—C6—C7—O2 | −26.5 (3) | N2—C5—C4—C3 | −0.1 (3) |
| N2—C6—C7—O3 | 154.55 (16) | C4—N1—C1—C2 | −0.1 (2) |
| C1—N1—C4—C3 | 0.2 (2) | N1—C1—C2—C3 | 0.0 (2) |
| C1—N1—C4—C5 | 179.57 (16) | C4—C3—C2—C1 | 0.1 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.88 | 1.93 | 2.782 (2) | 162. |
| N2—H2···O2ii | 0.88 | 2.09 | 2.9372 (19) | 161. |
Symmetry codes: (i) −x+2, −y, −z+1; (ii) −x+3/2, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CF2214).
References
- Banwell, M. G., Hamel, E., Hockless, D. C. R., Verdier-Pinard, P., Willis, A. C. & Wong, D. J. (2006). Bioorg. Med. Chem.14, 4627–4638. [DOI] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Bruker (1999). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Faulkner, D. J. (2002). Nat. Prod. Rep.18, 1–48. [DOI] [PubMed]
- Sheldrick, G. M. (1997). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sosa, A. C. B., Yakushijin, K. & Horne, D. A. (2002). J. Org. Chem.67, 4498–4500. [DOI] [PubMed]
- Zeng, X.-C. (2006). Acta Cryst. E62, o5505–o5507.
- Zeng, X.-C., Zeng, J., Li, X. & Ling, X. (2007). Acta Cryst. E63, o3424.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808027451/cf2214sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808027451/cf2214Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Enhanced figure: interactive version of Fig. 1




