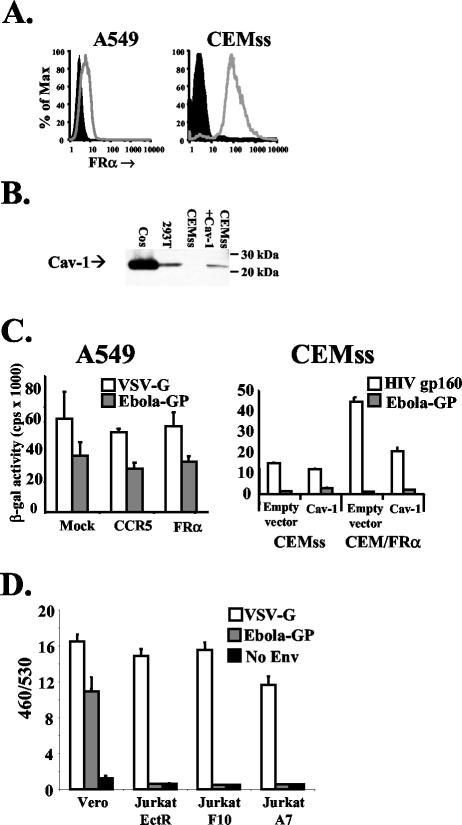
FIG. 2.
Expression of FRα and Cav-1 in cell lines does not enhance Ebola virus pseudotype infection. (A) Surface levels of FRα were assessed as described in the legend to Fig. 1 for A549 cells (left panel) transduced with lentiviral vectors (for protocols, see reference 31) expressing a control of CCR5 (filled histogram) or FRα (open histogram) and for CEMss cells (right panel, filled histogram) or CEM cells stably expressing FRα (open histogram). (B) Total protein levels of Cav-1 were assessed by Western blotting using a directly horseradish peroxidase-conjugated polyclonal rabbit antibody raised against the N terminus of Cav-1 (N-20; Santa Cruz Biotechnology, Santa Cruz, Calif.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed on cell lysates from 106 cells. CEMss cells were transduced with MLV amphotropic envelope pseudotyped MLV particles carrying the MIGR1 packaging vector encoding human CD8 (27) or MLV MIGR1 encoding human Cav-1. Cos and 293 cells were also tested to determine endogenous levels of Cav-1. (C) Mock-transduced and CCR5- or FRα-transduced A549 cells (left panel) or CEMss and CEM/FRα cells transduced with control or Cav-1-encoding MLV pseudovirions (right panel) were challenged with HIV-based pseudovirions encoding LacZ (18) pseudotyped with Ebola virus GP, VSV-G, or HIV gp160 (HXB2 strain). Two days postchallenge, cells were lysed and soluble β-galactosidase (β-gal) levels were measured with a chemiluminescence kit (Galactostar; Tropix) as per the manufacturer's instructions. Similar results were gained by using pseudovirions encoding luciferase as a reporter gene (data not shown). (D) Vero cells, but not Jurkat cells stably expressing ecotropic MLV receptor or previously described derivates expressing FRα (6), were transducible with VLPs bearing Ebola virus GP. VLPs were made by cotransfecting293T cells with eqimolar amounts of a Bla-VP40 fusion protein and GP, VSV-G, or empty vector. Forty-eight hours posttransfection, filtered supernatant was collected and diluted. Cells (5 × 105) were spin infected at 2,500 rpm for 2 h at 4°C and then incubated for 4 h at 37°C to allow entry. Cells were then labeled with CCF2-AM (Invitrogen, Carlsbad, Calif.) as per the manufacturer's instructions, washed, transferred to clear-bottomed, black-walled microtiter plates, and incubated overnight at room temperature to allow the enzymatic reaction to occur. The following morning, plates were read at 460 and 530 nm (Cytofluor 4000; Applied Biosystems, Foster City, Calif.). Background fluorescence from unlabeled cells was subtracted, and the ratio of fluorescence at 460 nm to that at 530 nm (460/530) was calculated for each well. Means and standard deviations shown are calculated from the results of four replicates, and the results presented are representative of three independent experiments.