Abstract
The origin of replication of African cassava mosaic virus (ACMV) and a gene expression vector based on Potato virus X were exploited to devise an in planta system for functional analysis of the geminivirus replication-associated protein (Rep) in transgenic Nicotiana benthamiana line pOri-2. This line contains an integrated copy of a tandem repeat of the ACMV origin of replication flanking nonviral sequences that can be mobilized and replicated by Rep as an episomal replicon. A Rep-GFP fusion protein can also mobilize and amplify the replicon, facilitating Rep detection in planta. The activity of Rep and its mutants, Rep-mediated host response, and the correlation between Rep intracellular localization and biological functions could be effectively assessed by using this in planta system. Our results indicate that modification of amino acid residues R2, R5, R7 and K11 or H56, L57 and H58 prevent Rep function in replication. This defect correlates with possible loss of Rep nuclear localization and inability to trigger the host defense mechanism resembling a hypersensitive response.
The genome of African cassava mosaic virus (ACMV; family Geminiviridae, genus Begomovirus) comprises two circular single-stranded DNA (ssDNA) components (DNA-A and DNA-B), both of which are required for systemic infection and symptom development in plants (45, 47). DNA-A can replicate autonomously (30, 50) and encodes six genes, two on the virion-sense strand and four on the complementary-sense strand, involved in rolling circle replication (43), transcriptional control of gene expression, the production of virus particles, and the induction of plant defense and viral counterdefense measures (11, 19, 20, 23, 25, 36, 49, 52, 55). DNA-B relies on DNA-A for replication and encodes two genes, one each on the virion-sense and complementary-sense strands, responsible for virus movement in plants (10, 15, 56, 57). DNA-A and DNA-B share an almost identical common region of about 200 nucleotides containing bidirectional promoters and the origin of replication (13, 23, 24, 46, 58).
The multifunctional replication-associated protein (Rep) participates in rolling circle replication and the control of viral and host gene expression (21, 33). Rep may also play an important role in the host response to infection. Thus, overexpression of ACMV Rep evokes a reaction resembling a hypersensitive response (HR), producing rapid local cell death and a systemic burst of H2O2 production in Nicotiana benthamiana (52). HR represents an active plant defense mechanism that is often elicited by virus-encoded proteins during the interplay between virus infection and plant defense (8, 17, 40). A range of plant RNA and DNA virus proteins involved in encapsidation, virus movement, and replication can act as HR elicitors (2, 6, 9, 16, 27-29, 34, 35, 39, 41, 44). Other viral proteins, including the begomovirus transcriptional activator protein (TrAP), are able to suppress the posttranscriptional gene silencing (PTGS) plant defense mechanism (53-55).
The functional analysis of Rep during the ACMV infection cycle is hampered by the complex interplay of viral protein expression and function and by the fact that Rep plays an essential role in this process and that mutations are often lethal. Hence, to assess the biological significance of the biochemical and molecular functions of Rep obtained from heterologous systems and in vitro assays in plants represents a major challenge. Here, we report a novel system in which the function of ACMV Rep in replication, its intracellular localization, and its role in the induction of the host response to infection can be simultaneously investigated.
MATERIALS AND METHODS
Clone construction.
The construction of pAV1Pro-GUS and pGUS, referred to here as pOri-1 and pOri-0, respectively, has been described (24). A fragment of the ACMV DNA-A common region, including the origin of replication, was PCR amplified by using primers CAATTGcAtgCACTCAACTAG and GATTGGCatgCATAAGTAGTG. Lowercase letters indicate changes introduced to create an SphI site (underlined). The PCR fragment was digested with SphI and cloned into the unique SphI site of pOri-1 to produce pOri-2, which contains a direct repeat of the origin of replication flanking the β-glucuronidase (GUS) coding region (Fig. 1A). The SstI/XhoI fragment of pOri-2 was cloned into the Agrobacterium tumefaciens binary vector pBin19 (3) to produce pBinOri-2.
FIG. 1.
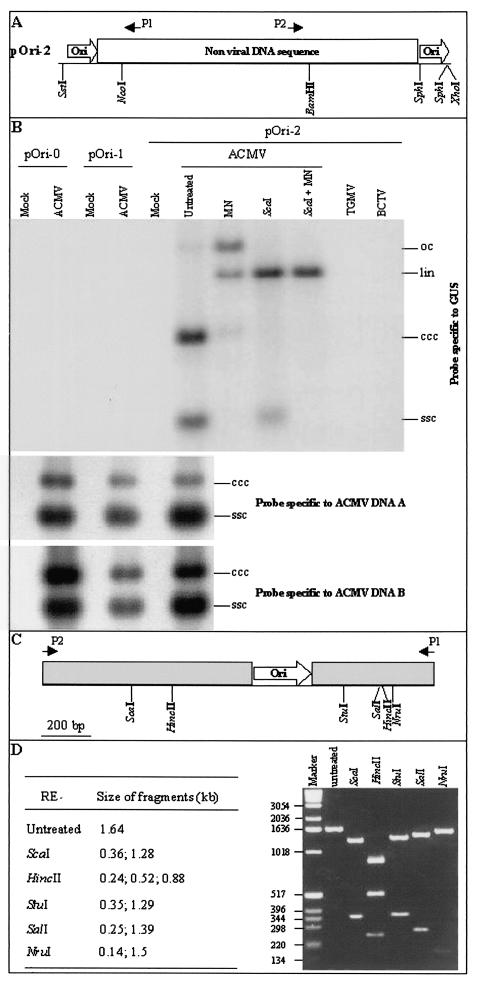
Detection of episomal replicon in ACMV-infected N. benthamiana line pOri-2. (A) The transgene contains a direct repeat of the ACMV replication origin (ori) flanking a nonviral DNA fragment including the β-glucuronidase (GUS) coding sequence and polyadenylation region. The positions of the GUS-specific primers, P1 and P2, are indicated. (B) DNA was extracted from N. benthamiana lines pOri-0, pOri-1, and pOri-2. Samples extracted from mock-inoculated plants and from plants infected with either ACMV, TGMV, or BCTV were either untreated or treated with mung bean nuclease (MN), ScaI (ScaI), or both (ScaI + MN). Blots were hybridized with probes specific to GUS, ACMV DNA-A, and ACMV DNA-B. The positions of circular ssDNA (ssc) and covalently closed circular (ccc), linear (lin), and open-circular (oc) dsDNA forms of the replicon and ACMV DNA components are indicated. (C) Structure of the predicted 1.64-kbp replicon fragment produced by PCR amplification by using GUS-specific primers P1 and P2. (D) Restriction endonuclease analysis of the 1.64-kbp PCR fragment. PCR was performed by using DNA extracted from ACMV-infected line pOri-2. The predicted sizes of DNA fragments generated from each digestion and the positions and sizes of DNA markers (bp) are indicated.
The construction of PVX/AC14, PVX/AC1m4, PVX/ACm1m4, and PVX/GFP has been described (51, 52). The green fluorescent protein (GFP) coding sequence, isolated from TXS.GFP-CP (42) as an EagI/BspEI fragment, was fused in frame to the Rep coding sequence in PVX/AC14 and PVX/AC1m4 to produce PVX/AC14-GFP and PVX/AC1m4-GFP, respectively. Fragments encompassing the Rep coding sequence were PCR amplified from PVX/AC1m4 for construction of AC1-GFP expression vectors (see Fig. 4A). The Rep coding sequence, containing alanine substitutions at residues R2, R5, R7 and K11, was amplified by using primers PP208 (GACTTGGgcgccATGgcAACTCCTgcTTTTgcAATTCAAGCCgcGAATGTCTTTCTC) and PP77 (52). The PCR product was digested with NarI and EagI and was cloned into the ClaI/EagI sites of PVX/GFP (51) to produce PVX/AC1(R2A-R5A-R7A—K11A)-GFP. Primers PP209 (GTTCTAATCCCgcgGAACACCTACTGTCA) and PP210 (GGTGTTCcgcGGGTATAGAACACTTTG) were designed to replace K24 with alanine, and primers PP211 (GAACCTgcCgcGgcTGCCCTCATTCAATTCGAG) and PP212 (GAGGGCAgcCgcGgcAGGTTCCCCATTCTG) were designed to substitute alanines for H56, L57, and H58. Lowercase letters in these primers indicate nucleotides modified to alter codons and to create a SacII site (underlined). The 5′ region of the Rep coding sequence was PCR amplified using either primers PP210 or PP212 together with PP50 (52), while the 3′ region was amplified using primers PP209 or PP211 together with PP77 (52). PCR products corresponding to the two halves of the coding sequences were digested with SacII and either NarI or EagI, and were cloned into the ClaI/EagI sites of PVX/GFP to produce PVX/AC1(K24A)-GFP and PVX/AC1(HLH58AAA)-GFP. The presence of the mutations in these constructs was confirmed by nucleotide sequencing.
FIG. 4.
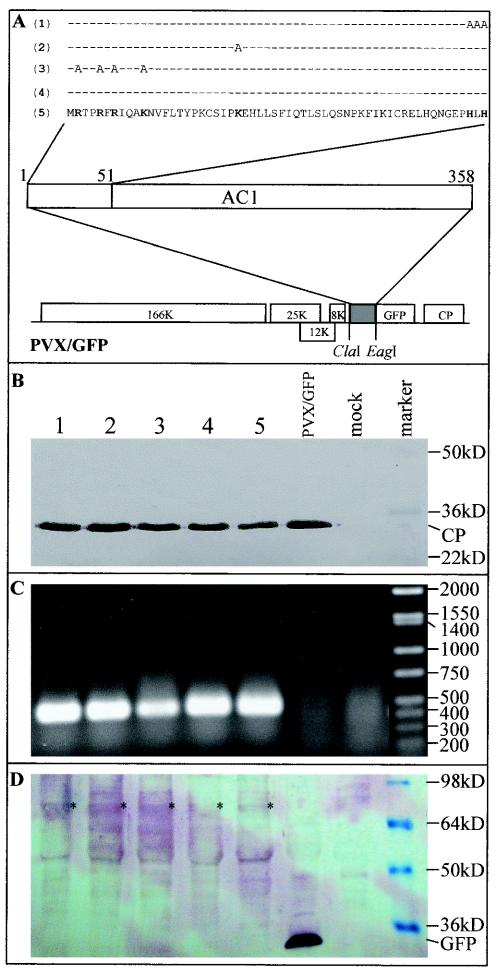
Expression Rep-GFP fusion protein mutants from PVX vectors. (A) Rep coding sequences were fused in frame to the GFP coding sequence in PVX/GFP vectors. The N-terminal 51 amino acids of Rep and the mutated amino acids within this region are indicated for PVX/AC14-GFP (5), PVX/AC1m4-GFP (4), PVX/AC1(R2A-R5A-R7A—K11A)-GFP (3), PVX/AC1(K24A)-GFP (2), and PVX/AC1(HLH58AAA)-GFP (1). (B) Western blot analysis of PXV coat protein (CP) expression. Proteins were extracted either from mock-inoculated plants (mock) or from plants infected with PVX/AC1(HLH58AAA)-GFP (lane 1), PVX/AC1(K24A)-GFP (lane 2), PVX/AC1(R2A-R5A-R7A—K11A)-GFP (lane 3), PVX/AC1m4-GFP (lane 4), PVX/AC14-GFP (lane 5), and PVX/GFP. The blot was probed with an antiserum raised against PVX coat protein. The positions and sizes of protein markers and the coat protein are indicated. (C) Detection of the Rep mRNA by RT-PCR. The positions and sizes of DNA markers (bp) are indicated. (D) Western blot analysis of Rep-GFP fusion protein expression. The blot was probed with an antiserum raised against GFP. The positions and sizes of protein markers, free GFP, and Rep-GFP fusion protein (*) are indicated. The samples in each lane of panels C and D correspond to those described in panel B.
Plant transformation.
Clone pBinOri-2 was mobilized into A. tumefaciens LBA4404 (22) by triparental mating (7), and transconjugants were selected for their resistance to kanamycin and rifampin. N. benthamiana was transformed with a leaf disk method (26), and transformants were regenerated following selection with kanamycin and carbencillin. Following self-fertilization, F1 and F2 progeny were tested for antibiotic sensitivity by germinating seeds on selective medium containing 0.5 mg of kanamycin/ml, and the ratio of the numbers of kanamycin-resistant and -sensitive plantlets was used to estimate the copy number of integrated DNA loci. Of five independent transgenic N. benthamiana lines, three contained a single copy of the pOri-2 transgene, one of which was chosen for further investigation. The production of transgenic N. benthamiana lines transformed with a single copy of either pOri-0 or pOri-1 has been described (24).
Plant inoculation and maintenance.
N. benthamiana plants were agroinoculated with infectious clones of ACMV (pBin1.3A and pBin2B), TGMV (pBincsTA1.6 and pBincsTB1.4), and BCTV (pBin1.2) as described earlier (4, 31, 56). Alternatively, RNA transcripts were produced by in vitro transcription of the recombinant PVX constructs after linearization with SpeI and were mechanically inoculated onto N. benthamiana plants as described (5). Plants were maintained in an insect-free containment greenhouse or growth room at 25°C with supplementary lighting to give a 12-h photoperiod. Local and systemic symptom development was assessed on a daily basis and was photographically recorded with a Nikon Digital Camera Coolpix 995.
Detection and characterization of replicon DNA.
DNA was extracted from leaves systemically infected either with ACMV, TGMV, or BCTV at 10 days postinoculation (dpi) with a DNeasy Plant Mini Kit (Qiagen). DNA aliquots (5 μg) were treated with mung bean nuclease to remove ssDNA and were digested with ScaI. DNA samples were resolved on a 1.4% agarose gel in TNE buffer (40 mM Tris-acetate, pH 7.5, 20 mM sodium acetate, 2 mM EDTA), transferred to nylon membrane and detected by hybridization with radiolabeled probes specific to either ACMV DNA-A, ACMV DNA-B, or transgene sequences.
A PCR-based approach was developed to detect replicon mobilization and replication by use of primers P1 (TCGCGCTGATACCAGACGTTGC) and P2 (GGACTGGCATGAACTTCGGTG) that are specific for the GUS coding sequence in the transgene (Fig. 1A). Restriction endonuclease analysis and DNA sequencing were performed to verify the integrity of the PCR fragments.
To detect the ACMV fragments in the recombinant PVX constructs and to investigate whether replicon replication had occurred in PVX-infected plants, nucleic acids were extracted from leaf tissues at 7 dpi by using a DNeasy Plant Mini Kit (Qiagen) according to the manufacturer's protocol, with the omission of RNase treatment. Reverse transcriptase PCR (RT-PCR) was performed with primers PP95 and PP96 as previously described (52). Detection of the replicon was carried out by PCR amplification and Southern blot hybridization with a digoxigenin (DIG)-labeled replicon-specific probe prepared with a DIG DNA labeling and detection kit (Roche).
Analysis of AC1-GFP fusion protein expression.
To investigate PVX coat protein and AC1-GFP fusion protein expression in plants, proteins were extracted from leaf tissues as described by Hong et al. (24). Western blot analyses of protein aliquots (10 μg) were performed with a polyclonal antiserum raised against PVX coat protein or GFP and were detected by using goat anti-rabbit immunoglobulin G conjugated with alkaline phosphatase (Sigma) and 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrates (Roche).
Subcellular localization of AC1-GFP fusion protein in plant cells.
N. benthamiana leaves infected with either PVX/GFP, PVX/AC14-GFP, PVX/AC1m4-GFP, PVX/AC1(R2A-R5A-R7A—K11A)-GFP, PVX/AC1(K24A)-GFP, or PVX/AC1(HLH58AAA)-GFP were cut into 3-mm-wide strips, vacuum infiltrated, and fixed overnight in 4% paraformaldehyde, 100 mM phosphate buffer (pH 7.0). Tissues were then infiltrated with 15% sucrose-100 mM phosphate buffer (pH 7.0), embedded in 5% low-melting-point agarose, and sectioned in a cryostat at −20°C (Bright Instruments OTS). Sections (10 μm) were mounted in 50% glycerol containing 1 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml and were examined by using a Zeiss Axiophot microscope equipped with a Nikon Digital Camera Coolpix 995. Fluorescence was observed with filters for GFP (450- to 490-nm excitation, 520-nm long-pass emission) and DAPI (365-nm excitation, 420-nm long-pass emission).
RESULTS
Mobilization and amplification of the pOri-2 replicon during ACMV infection.
The common region of ACMV, containing the origin of replication, was isolated from the DNA-A component. Transgenic line pOri-2 was produced by transforming N. benthamiana with a single copy of a fragment containing a direct repeat of the common region flanking the β-glucuronidase (GUS) coding region (Fig. 1A). Lines pOri-0 and pOri-1 (24) contain single-copy inserts of the GUS coding sequence alone and a single copy of the origin of replication located upstream of the GUS coding sequence, respectively. All plant lines were susceptible to infection by ACMV as well as isolates of Tomato golden mosaic virus (TGMV) and Beet curly top virus (BCTV), and plants developed typical systemic symptoms associated with each virus infection. Although ACMV DNA-A and DNA-B components were readily detectable in infected plant lines, the transgene episomal DNA (referred to here as the replicon) were present only in line pOri-2 (Fig. 1B). The presence of both single- and double-stranded (ds) DNA forms of the replicon was confirmed by mung bean nuclease digestion that degraded the ssDNA and nicked the covalently closed circular dsDNA to produce linear and open-circular dsDNA forms. Digestion with ScaI, a single cutting restriction endonuclease within the GUS coding region of the transgene, produced linear dsDNA of approximately 3.2 kbp, the anticipated size of a mobilized version of the transgene. In contrast to ACMV infection, neither TGMV nor BCTV was able to mobilize the ACMV-based replicon in line pOri-2 infected with these viruses (Fig. 1B).
To confirm the presence of circular replicon DNA in ACMV-infected line pOri-2, GUS-specific primers P1 and P2 (Fig. 1A) were used for PCR amplification of transgene sequences. Their divergent orientation within the GUS coding region ensured that the primers only amplified a fragment from a circular derivative of the transgene, not from the linear integrated sequence. A PCR fragment with a predicted size of 1.64 kbp (Fig. 1C) was amplified with these primers and DNA samples from ACMV-infected line pOri-2. No product was amplified from mock-inoculated line pOri-2 and ACMV-infected lines pOri-0 and pOri-1. The integrity of the replicon fragment was confirmed by restriction endonuclease digestion of the 1.64-kbp fragment (Fig. 1D), and sequencing analysis confirmed that the fragment contained the intact GUS coding region and only a single copy of the ACMV origin of replication.
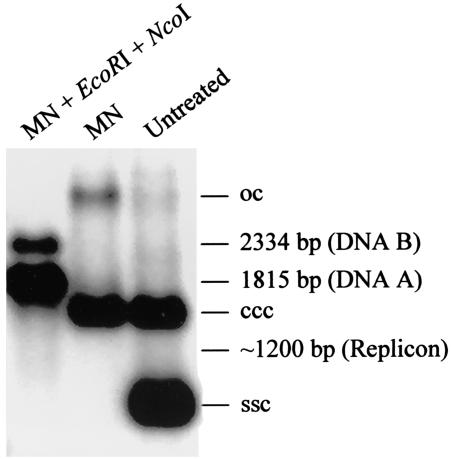
To address the efficiency of replicon replication, the levels of replicon DNA versus ACMV A and B DNAs in ACMV-infected line pOri-2 plants were compared. This was achieved by Southern blot analysis with 32P-labeled probes specific to the sequence of replication origin that specified DNAs corresponding to either the replicon or ACMV DNA-A and B (Fig. 2). As predicted from digestion of total DNAs extracted from ACMV-infected line pOri-2 plant with restriction endonucleases EcoRI and NcoI plus mung bean nuclease, a 1.815-kbp A- and a 2.334-kbp B-specific DNA fragment were readily detected. However, an approximately 1.2-kbp replicon-specific fragment was only vaguely seen in the same blot. These data indicated that only a very low level of replicon DNA accumulated when compared to that of ACMV genomes in the ACMV-infected line pOri-2 plant.
FIG. 2.
Comparisons of the levels of pOri-2 replicon versus ACMV A and B DNAs. Total DNAs extracted from N. benthamiana lines pOri-2 were either untreated or treated with mung bean nuclease (MN) or MN with EcoRI and NcoI (MN + EcoRI + NcoI). Blots were hybridized with probes specific to the ACMV replication origin. The positions of circular ssDNA (ssc) and covalently closed circular (ccc), and open-circular (oc) dsDNAs are indicated. The predicted sizes and positions of ACMV A- and B- and replicon-specific DNA fragments generated from the treatment are indicated.
Taken together, our data are consistent with Rep-mediated nicking at one origin in the transgene and with nicking and ligation of the displaced nascent ssDNA at the second origin after replication of the transgene sequence. In this way, a circular episomal replicon is produced that is subsequently amplified by Rep and the replication enhancer protein (REn) provided in trans from the ACMV genome. The highly specific interaction of Rep with its cognate origin of replication is conferred by short reiterated motifs (21). Consistent with this, neither TGMV nor BCTV is able to mobilize the ACMV replicon because their Reps recognize distinct motifs.
Analysis of Rep activity when expressed in line pOri-2 from a PVX vector.
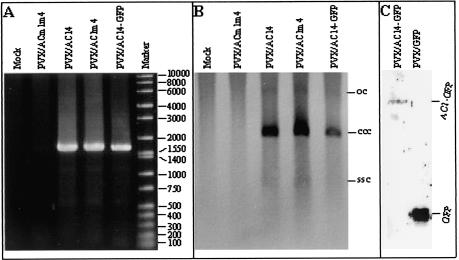
Previously, we have demonstrated that ACMV Rep can be expressed in N. benthamiana from a PVX vector (52). To investigate whether Rep expressed in this way is functionally active, RNA transcripts were produced in vitro from PVX/AC14, PVX/AC1m4, and PVX/ACm1m4 and were mechanically inoculated onto line pOri-2. By use of the PCR assay with primers P1 and P2, episomal copies of the replicon were detected in plants infected with PVX/AC14 (expressing both Rep and AC4 protein) and PVX/AC1m4 (expressing Rep alone) but not in plants infected with PVX/ACm1m4 (in which the expression of both Rep and AC4 protein had been disrupted) (Fig. 3A). Moreover, the level of covalently closed circular dsDNA was evident by Southern blot analysis in plants infected with either PVX/AC14 or PVX/AC1m4 but not PVX/ACm1m4 (Fig. 3B), although the ssDNA and open circular forms of the replicon were very low. Episomal replicon was also present in plants infected with PVX/AC14-GFP that can express a Rep-GFP fusion protein of approximately 67 kDa (Fig. 3A and B). Expression of the fusion protein from PVX/AC14-GFP, as well as GFP (26.9 kDa) from PVX/GFP, was confirmed by Western blot analysis using an antiserum raised against GFP (Fig. 3C).
FIG. 3.
Detection of episomal replicon in N. benthamiana line pOri-2 plants infected with PVX expression vectors. (A) DNA samples were extracted from line pOri-2 plants infected either with PVX/ACm1m4, PVX/AC14, PVX/AC1m4, or PVX/AC14-GFP and were used for PCR amplification of a fragment of the replicon with primers P1 and P2. The positions and sizes of DNA markers (bp) are indicated. (B) The same DNA samples were analyzed by Southern blotting with a GUS-specific probe to detect episomal replicon. The positions of circular single-stranded DNA (ssc) and covalently closed circular (ccc), and open circular (oc) dsDNA forms of the replicon are indicated. (C) Proteins were extracted from line pOri-2 plants infected with either PVX/AC14-GFP or PVX/GFP, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and detected by Western blot analysis by using antiserum raised against GFP. The positions of AC1-GFP and free GFP are indicated.
These data indicate that ACMV Rep expressed from the PVX vector is functionally active in mobilizing and amplifying the replicon from the transgene in line pOri-2 although the PVX system lacks an REn AC3. Moreover, Rep containing a functional GFP tag at its C terminus retains this activity. When the replicon ss- and dsDNA profile is taken into account, it is unlikely that such a lack of REn, which interacts with Rep and is required for efficient begomovirus replication, may have a diminished effect on monitoring Rep replication activity. It is also worth noting that the production of Rep in plants infected with begomoviruses, including ACMV, is tightly regulated and balanced for the benefit of the viral life cycle in order to carry out the function of viral DNA replication. On the other hand, the level of Rep is expected to be higher in the PVX-based system. This indeed reflects the readiness of nonradioactive Southern detection of replicon, indicating that replicon replication may occur more efficiently in line pOri-2 plants when Rep is expressed from the PVX vector than from ACMV DNA-A.
Expression and analysis of Rep-GFP mutants in line pOri-2.
To explore the potential of line pOri-2 as an indicator of Rep function, Rep mutants were fused in frame to GFP and were expressed in plants from PVX vectors (Fig. 4A). GFP-tagged Rep mutants had alanine substitutions at positions R2, R5, R7 and K11 in PVX/AC1(R2A-R5A-R7A—K11A)-GFP, K24 in PVX/AC1(K24A)-GFP and H56, L57 and H58 in PVX/AC1(HLH58AAA)-GFP. It should be noted that none of these vectors is able to express AC4 protein. All of the vectors were infectious in plants, and Western blot analysis indicated that the PVX coat protein (27 kDa) accumulated to approximately similar levels in each case (Fig. 4B). In addition, RT-PCR analysis with primers specific for the Rep coding region (PP95 and PP96) produced the predicted 430-bp fragment (Fig. 4C), suggesting that the Rep coding sequences were stably maintained in the vector during systemic infection of the plant. Moreover, a nearly same level of the different mutant Rep proteins was detected by immunoblotting in plants infected with each of the individual recombinant PVXs (Fig. 4D), indicating that the mutations did not destabilize these Rep-GFP fusion proteins.
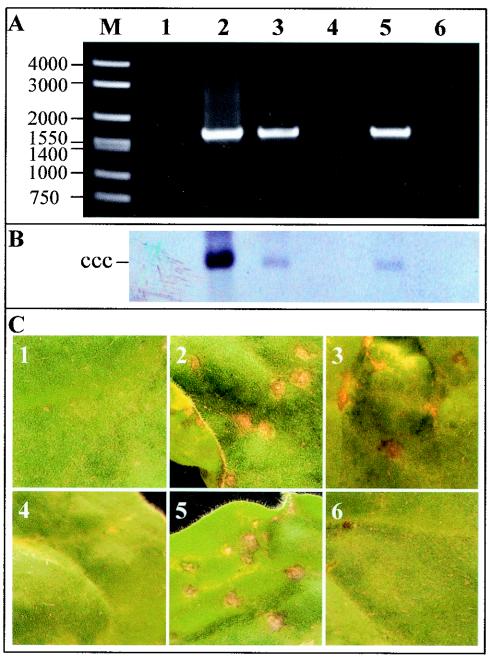
The Rep-GFP fusion protein expressed from both PVX/AC14-GFP and PVXAC1m4-GFP was able to mobilize the replicon in line pOri-2 to produce the diagnostic 1.64-kbp fragment using the PCR assay with primers P1 and P2 (Fig. 5A, lanes 2 and 3). The PCR product was also obtained in plants infected with PVX/AC1(K24A)-GFP (lane 5) but not with either PVX/AC1(R2A-R5A-R7A—K11A)-GFP or PVX/AC1(HLH58AAA)-GFP (lanes 4 and 6). These data were consistent with Southern blot detection of replicon replication (Fig. 5B). It should be noted that a relatively higher level of replicon DNA was detected in plants infected with PVX/AC14-GFP. Nevertheless, the similar amounts of the covalently closed circular dsDNA of the replicon accumulated in plants infected with PVXAC1m4-GFP and PVX/AC1(K24A)-GFP, respectively, indicated that the K24A mutation had no obvious impacts on the efficiency of Rep function. The biological activity of Rep-GFP fusion protein mutants was further assessed by examining the phenotypes associated with each infection in line pOri-2. Plants challenged with either PVX/AC14-GFP, PVX/AC1m4-GFP, or PVX/AC1(K24A)-GFP developed local and systemic symptoms identical to those previously described for PVX/AC14 and PVX/AC1m4 (52). The production of necrotic lesions resembling HR appeared on inoculated leaves by 3 to 5 dpi (Fig. 5C, panels 2, 3, and 5). PVX/AC14-GFP infection produced severe systemic necrosis by 6 or 7 dpi, causing the collapse of young developing tissues, while PVX/AC1m4-GFP and PVX/AC1(K24A)-GFP infection produced only veinal chlorosis on young leaves. Plants infected with either PVX/AC1(R2A-R5A-R7A—K11A)-GFP or PVX/AC1(HLH58AAA)-GFP developed chlorotic lesions on inoculated leaves (Fig. 5C, panels 4 and 6) and systemic chlorosis on young leaves typical of PVX and PVX/GFP infection (Fig. 5C, panel 1).
FIG. 5.
Effect of Rep mutations on replicon mobilization and phenotype in N. benthamiana line Ori-2 plants. (A) DNA samples were extracted from plants infected with PVX/GFP (lane 1), PVX/AC14-GFP (lane 2), PVX/AC1m4-GFP (lane 3), PVX/AC1(R2A-R5A-R7A—K11A)-GFP (lane 4), PVX/AC1(K24A)-GFP (lane 5), and PVX/AC1(HLH58AAA)-GFP (lane 6) and were used for PCR amplification of a fragment of the replicon with primers P1 and P2. The positions and sizes of DNA markers (bp) are indicated. (B) Southern blot analysis of replicon replication. The samples in each lane correspond to those described for panel A. The position of the covalently closed circular (ccc) DNA form of the replicon is indicated. (C) Symptoms induced in plants infected with PVX vectors expressing Rep-GFP fusion protein mutants. Leaves show local necrosis when inoculated with PVX/AC14-GFP (panel 2), PVX/AC1m4-GFP (panel 3), and PVX/AC1(K24A)-GFP (panel 5) but show chlorotic lesions when inoculated with PVX/GFP (panel 1), PVX/AC1(R2A-R5A-R7A—K11A)-GFP (panel 4), and PVX/AC1(HLH58AAA)-GFP (panel 6). Leaves were photographed at 5 dpi.
Subcellular localization of Rep mutants.
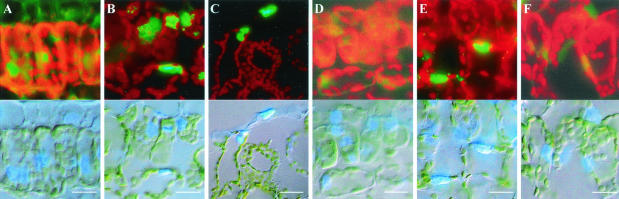
The biological functions and phenotypes of the Rep mutants were correlated with their cellular localization. Young leaf tissue from N. benthamiana plants infected with the PVX vectors was collected and examined by fluorescence microscopy. Representative sections of leaf mesophyll cells are shown in Fig. 6. As reported previously, GFP fluorescence was present throughout the cytoplasm of cells infected with PVX/GFP (panel A). Fluorescence was localized predominantly in the nuclei of cells infected with either PVX/AC14-GFP, PVX/AC1m4-GFP or PVX/AC1(K24A)-GFP (panels B, C and E) although some fluorescence was occasionally observed in the cytoplasm of some cells. In contrast, GFP fluorescence was not particularly restricted to the nuclei of cells infected with either PVX/AC1(R2A-R5A-R7A—K11A)-GFP or PVX/AC1(HLH58AAA)-GFP, but occurred throughout the cytoplasm (panels D and F). These data suggest that residue K24 has little effect on Rep nuclear localization, while one or more of the N-terminal residues R2, R5, R7, and K11, as well as H56, L57 and H58, are essential for this process.
FIG. 6.
Subcellular localization of Rep-GFP fusion protein mutants. Leaf tissues infected with PVX/GFP (A), PVX/AC14-GFP (B), PVX/AC1m4-GFP (C), PVX/AC1(R2A-R5A-R7A—K11A)-GFP (D), PVX/AC1(K24A)-GFP (E), and PVX/AC1(HLH58AAA)-GFP (F) were screened for fluorescence by using filters for GFP (450- to 490-nm excitation and 520-nm long-pass emission) (top panels) and DAPI (365-nm-long excitation and 420-nm long-pass emission) under light-field illumination (bottom panels). Chloroplast autofluorescence appears red. Bar = 10 μm.
DISCUSSION
Geminiviruses have small circular DNA genomes with overlapping genes and a complex intergenic region containing bidirectional promoter elements interspersed within the origin of replication. Rep is the only begomovirus protein that is indispensable for viral DNA replication, and mutations introduced into the protein are frequently lethal to the virus. It initiates rolling circle replication by binding to specific motifs (iterons) within the origin of replication (1, 12) and by introducing a nick in the virion-sense strand within a highly conserved TAATATT↓AC motif (46). Because the iterons are located downstream of the complementary-sense promoter, Rep binding serves to down-regulate its own expression and probably that of the overlapping AC4 protein (18, 19, 23, 48). Rep also plays a role in modulating the cell cycle to provide conditions suitable for viral DNA replication in differentiated cells (32, 37), and its expression induces a host defense response that may be countered by AC4 protein (52). The participation in such a complex interplay of processes precludes the detailed analysis of Rep functions by screening virus mutants for their ability to infect plants. To overcome this problem, we have developed a convenient assay in which ACMV Rep is introduced and expressed from a PVX-based vector, and its function in viral DNA replication is assessed by screening for the mobilization and amplification of a 3.2-kbp circular replicon from an integrated transgene containing a nonviral fragment (GUS coding sequences) flanked by copies of the ACMV origin of replication. The assay was refined by fusing GFP to the C terminus of Rep, providing a simple nondestructive method for Rep detection. We have found that the Rep-GFP fusion has properties similar to those of Rep with respect to viral DNA replication, cell localization, and host response.
The replicon was mobilized and amplified when Rep was expressed from either the ACMV genome or from a PVX vector, and both single-stranded and double-stranded DNA forms of the replicon were produced, indicative of rolling circle replication. The replicon was mobilized only when flanking copies of the origin were present on the integrated linear template, mimicking a circular template with a single origin. Thus, Rep-mediated initiation, termination of replication at two different transgenic origins, and circularization of the nascent strand will produce a replicon containing a single intact copy of the origin. A previous investigation with a similar construct based on an ACMV DNA-A deletion mutant with flanking origins of replication failed to mobilize a 1,900-bp replicon in this way (14). The reason for this different behaviour is unclear but presumably resides in the difference in composition or size of the transgenes that could affect the mobilization, replication, and/or stability of the replicons.
We have used this assay to screen selected Rep alanine-scanning mutants for their biological activity. These amino acid residues were chosen for mutagenesis because they were highly conserved among the Reps of many monopartite and bipartite begomoviruses, and likely involved in targeting Rep to nuclei. Substitution of lysine at position 24 had no effect on the ability of Rep to mobilize and replicate the replicon, and it did not alter either Rep nuclear localization or the host response to Rep overexpression. In contrast, substitution of four N-terminal basic amino acids (arginines at positions 2, 5 and 7 and lysine at position 11) prevented mobilization of the replicon. The mutant was no longer specifically localized within the nucleus and did not trigger host response. This implies that a nuclear localization signal had been disrupted, thereby probably effectively excluding Rep from the nucleus, where it is normally active. However, because GFP localizes to cytoplasm and nuclei in plant cells in its native form, we cannot rule out that traces of the mutated Rep-GFP may still diffuse to nuclei. In this scenario, inefficient nuclear import and other deficit effects on the Rep by such mutations may account for “loss of function” of the Rep mutant. The impact of each individual mutation at R2A, R5A, R7A, and K11A on nuclear localization and biological functions of the ACMV Rep remains to be elucidated. Finally, substitution of histidine-leucine-histidine (positions 56 to 58), a motif that is conserved in many initiator proteins involved in rolling circle replication, also prevented mobilization of the replicon. This is consistent with mutagenesis of the TGMV Rep motif that prevented viral DNA cleavage and replication (21, 38). In addition, we have demonstrated that the mutant is no longer nuclear localized, providing an explanation for its inability to mobilize and amplify the replicon in planta.
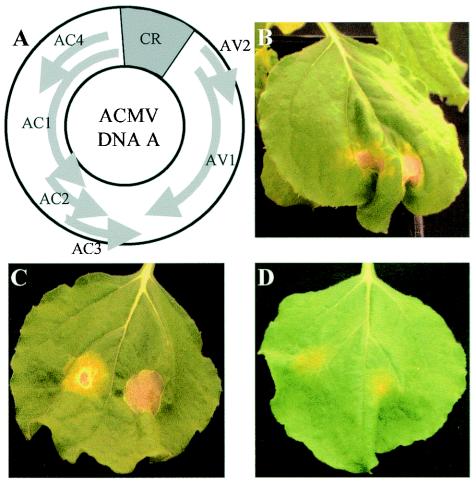
Expression of ACMV Rep either from a PVX vector or by agroinfiltration elicited a response resembling HR: the induction of local necrosis and a systemic burst of H2O2 production in N. benthamiana (52). Moreover, agroinfiltration of N. benthamiana with ACMV DNA-A or together with DNA-B also triggered a local necrotic response, while agroinfiltration with ACMV DNA-B alone induced no such a response (Fig. 7). Our present findings are consistent with Rep being responsible for the induction of the host response and demonstrate that the two Rep mutants that are dysfunctional in replication and nonspecific nuclear localization are also unable to induce this response.
FIG. 7.
ACMV DNA-A-mediated induction of local necrosis response in N. benthamiana. (A) Genome organization of ACMV DNA-A. (B to D) Plants were infiltrated with A. tumefaciens LBA4404 carrying pBin1.3A for DNA-A (B), pBin2B for DNA-B (D), and both (C). Necrosis was induced only in plants after agroinfiltration with ACMV DNA-A alone or with DNA-B but not DNA-B alone. Leaves were photographed 20 days postagroinfiltration.
Acknowledgments
We thank D.C. Baulcombe for providing the original PVX gene expression vector and S. Santa Cruz for providing plasmid TXS.GFP-CP and antibodies against GFP and PVX coat protein. We are grateful to T. M. A. Wilson for his encouragement throughout the work.
This project was supported by a BBSRC CSG grant to Y.H. and a grant from the BBSRC Plant Molecular Biology Programme II to J.S.
REFERENCES
- 1.Argüello-Astorga, G. R., R. G. Guevara-González, L. R. Herrera-Estrella, and R. F. Rivera-Bustamante. 1994. Geminivirus replication origins have a group-specific organization of iterative elements: a model for replication. Virology 203:90-100. [DOI] [PubMed] [Google Scholar]
- 2.Berzal-Herranz, A., A. De La Cruz, F. Tenllado, J. R. Diaz-Ruiz, L. Lopez, A. I. Sanz, C. Vaquero, M. T. Serra, and I. Garcia-Luque. 1994. The Capsicum L3 gene-mediated resistance against tobamoviruses is elicited by the coat protein. Virology 209:498-505. [DOI] [PubMed] [Google Scholar]
- 3.Bevan, M. 1984. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12:8711-8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briddon, R. W., J. Watts, P. G. Markham, and J. Stanley. 1989. The coat protein of beet curly top virus is essential for infectivity. Virology 172:628-633. [DOI] [PubMed] [Google Scholar]
- 5.Chapman, S., T. Kavanagh, and D. Baulcombe. 1992. Potato virus X as a vector for gene expression in plants. Plant J. 2:549-557. [DOI] [PubMed] [Google Scholar]
- 6.Chu, M., J.-K. Park, and H. B. Scholthof. 1999. Separate regions on the tomato bushy stunt virus p22 protein mediate cell-to-cell movement versus elicitation of effective resistance responses. Mol. Plant-Microbe Interact. 12:285-292. [Google Scholar]
- 7.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon, R. A., M. J. Harrison, and C. J. Lamb. 1994. Early events in the activation of plant defence responses. Annu. Rev. Phytopathol. 32:479-501. [Google Scholar]
- 9.Erickson, F. L., S. Holzberg, A. Calderon-Urrea, V. Handley, M. Axtell, C. Corr, and B. Baker. 1999. The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18:67-75. [DOI] [PubMed] [Google Scholar]
- 10.Etessami, P., R. Callis, S. Ellwood, and J. Stanley. 1988. Delimitation of essential genes of cassava latent virus DNA-2. Nucleic Acids Res. 16:4811-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etessami, P., K. Saunders, J. Watts, and J. Stanley. 1991. Mutational analysis of complementary-sense genes of African cassava mosaic virus DNA A. J. Gen. Virol. 72:1005-1012. [DOI] [PubMed] [Google Scholar]
- 12.Fontes, E. P. B., H. J. Gladfelter, P. A. Eagle, P. S. Sipe, V. A. Luckow, and L. Hanley-Bowdoin. 1994. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 269:8459-8465. [PubMed] [Google Scholar]
- 13.Frey, P. M., N. G. Scharer-Hernandez, J. Futterer, I. Potrykus, and J. Puonti-Kaerlas. 2001. Simultaneous analysis of the bidirectional African cassava mosaic virus promoter activity using two different luciferase genes. Virus Genes 22:231-242. [DOI] [PubMed] [Google Scholar]
- 14.Frischmuth, T., and J. Stanley. 1998. Recombination between viral DNA and the transgenic coat protein gene of African cassava mosaic geminivirus. J. Gen. Virol. 79:1265-1271. [DOI] [PubMed] [Google Scholar]
- 15.Frischmuth, T., S. Roberts, A. von Arnim, and J. Stanley. 1993. Specificity of bipartite geminivirus movement proteins. Virology 196:666-673. [DOI] [PubMed] [Google Scholar]
- 16.Garrido-Ramirez, E. R., M. R. Sudarshana, W. J. Lucas, and R. L. Gilbertson. 2000. Bean dwarf mosaic virus BV1 protein is a determinant of the hypersensitive response and avirulence in Phaseolus vulgaris. Mol. Plant-Microbe Interact. 13:1184-1194. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg, J. T. 1997. Programmed cell death in plant-pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:525-545. [DOI] [PubMed] [Google Scholar]
- 18.Gröning, B. R., R. J. Hayes, and K. W. Buck. 1994. Simultaneous regulation of tomato golden mosaic virus coat protein and AL1 gene expression: expression of the AL4 gene may contribute to suppression of the AL1 gene. J. Gen. Virol. 75:721-726. [DOI] [PubMed] [Google Scholar]
- 19.Haley, A., X. Zhan, K. Richardson, K. Head, and B. Morris. 1992. Regulation of the activities of African cassava mosaic virus promoters by the AC1, AC2, and AC3 gene products. Virology 188:905-909. [DOI] [PubMed] [Google Scholar]
- 20.Haley, A., K. Richardson, X. C. Zhanm, and B. Morris. 1995. Mutagenesis of the BC1 and BV1 genes of African cassava mosaic virus identifies conserved amino acids that are essential for spread. J. Gen. Virol. 76:1291-1298. [DOI] [PubMed] [Google Scholar]
- 21.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18:1-106. [PubMed] [Google Scholar]
- 22.Hoekema, A., P. R. Hirsch, P. J. J. Hooykaas, and R. A. Schilperoort. 1983. A binary vector strategy based on separation of vir- and T-regions of the Agrobacterium tumefaciens Ti-plasmid. Nature 303:179-180. [Google Scholar]
- 23.Hong, Y., and J. Stanley. 1995. Regulation of African cassava mosaic virus complementary-sense gene expression by N-terminal sequences of the replication-associated protein AC1. J. Gen. Virol. 76:2415-2422. [DOI] [PubMed] [Google Scholar]
- 24.Hong, Y., K. Saunders, M. R. Hartley, and J. Stanley. 1996. Resistance of geminivirus infection by virus-induced expression of dianthin in transgenic plants. Virology 220:119-127. [DOI] [PubMed] [Google Scholar]
- 25.Hong, Y., K. Saunders, and J. Stanley. 1997. Transactivation of dianthin transgene expression by African cassava mosaic virus AC2. Virology 228:383-387. [DOI] [PubMed] [Google Scholar]
- 26.Horsch, R. B., and H. J. Klee. 1986. Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: role of the T-DNA borders in the transfer process. Proc. Natl. Acad. Sci. USA 83:4428-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kavanaugh, T., M. Goulden, S. Santa Cruz, S. Chapman, I. Barker, and D. Baulcombe. 1992. Molecular analysis of a resistance-breaking strain of potato virus X. Virology 189:609-617. [DOI] [PubMed] [Google Scholar]
- 28.Kim, C.-H., and P. Palukaitis. 1997. The plant defence response to cucumber mosaic virus in cowpea is elicited by the viral polymerase gene and affects virus accumulation in single cells. EMBO J. 16:4060-4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiraly, L., A. B. Cole, J. E. Bourque, and J. E. Schoelz. 1999. Systemic cell death is elicited by the interaction of a single gene in Nicotiana clevelandii and gene VI of cauliflower mosaic virus. Mol. Plant-Microbe Interact. 12:919-925. [Google Scholar]
- 30.Klinkenberg, F. A., and J. Stanley. 1990. Encapsidation and spread of African cassava mosaic virus DNA A in the absence of DNA B when agroinoculated to Nicotiana benthamiana. J. Gen. Virol. 71:1409-1412. [Google Scholar]
- 31.Klinkenberg, F. A., S. Ellwood, and J. Stanley. 1989. Fate of African cassava mosaic virus coat protein deletion mutants after agroinoculation. J. Gen. Virol. 70:1837-1844. [DOI] [PubMed] [Google Scholar]
- 32.Kong, L.-J., B. M. Orozco, J. R. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laufs, J., I. Jupin, C. David, S. Schumacher, F. Heyraud-Nitschke, and B. Gronenborn. 1995. Geminivirus replication: genetic and biochemical characterization of Rep protein function, a review. Biochimie 77:765-773. [DOI] [PubMed] [Google Scholar]
- 34.Malcuit, I., M. R. Marano, T. Kavanaugh, W. De Jong, A. Forsyth, and D. C. Baulcombe. 1999. The 25-kDa movement protein of PVX elicits Nb-mediated hypersensitive cell death in potato. Mol. Plant-Microbe Interact. 12:536-543. [Google Scholar]
- 35.Meshi, T., F. Motoyoshi, T. Maeda, S. Yoshiwoka, H. Watanabe, and Y. Okada. 1989. Mutations in the tobacco mosaic virus 30-kD protein gene overcome Tm-2 resistance in tomato. Plant Cell 1:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris, B., K. Richardson, P. Eddy, X. C. Zhan, A. Haley, and R. Gardner. 1991. Mutagenesis of the AC3 open reading frame of African cassava mosaic virus DNA-A reduces DNA-B replication and ameliorates disease symptoms. J. Gen. Virol. 72:1205-1213. [DOI] [PubMed] [Google Scholar]
- 37.Nagar, S., T. J. Pedersen, K. Carrick, L. Hanley-Bowdoin, and D. Robertson. 1995. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7:705-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orozco, B. M., and L. Hanley-Bowdoin. 1998. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem. 273:24448-24456. [DOI] [PubMed] [Google Scholar]
- 39.Padgett, H. S., and R. N. Beachy. 1993. Analysis of tobacco mosaic virus strain capable of overcoming N gene-mediated resistance. Plant Cell 5:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryals, J. A., U. H. Neuenschwander, M. G. Willits, A. Molina, H. Y. Steiner, and M. D. Hunt. 1996. Systemic acquired resistance. Plant Cell 8:1809-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito, T., T. Meshi, N. Takamatsu, and Y. Okada. 1987. Coat protein gene sequence of tobacco mosaic virus encodes a host response determinant. Proc. Natl. Acad. Sci. USA 84:6074-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santa Cruz, S., S. Chapman, A. G. Roberts, I. M. Roberts, D. A. Prior, and K. J. Oparka. 1996. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc. Natl. Acad. Sci. USA 93:6286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saunders, K., A. Lucy, and J. Stanley. 1991. DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res. 19:2325-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scholthof, H. B., K.-B. G. Scholthof, and A. O. Jackson. 1995. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7:1157-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanley, J. 1983. Infectivity of the cloned geminivirus genome requires sequences from both DNAs. Nature 305:643-645. [Google Scholar]
- 46.Stanley, J. 1995. Analysis of African cassava mosaic-virus recombinants suggests strand nicking occurs within the conserved nonanucleotide motif during the initiation of rolling circle DNA-replication. Virology 206:707-712. [DOI] [PubMed] [Google Scholar]
- 47.Stanley, J., and M. R. Gay. 1983. Nucleotide sequence of cassava latent virus DNA. Nature 301:260-262. [Google Scholar]
- 48.Sunter, G., M. D. Hartitz, and D. M. Bisaro. 1993. Tomato golden mosaic virus leftward gene expression: autoregulation of geminivirus replication protein. Virology 195:275-280. [DOI] [PubMed] [Google Scholar]
- 49.Townsend, R., J. Stanley, S. J. Curson, and M. N. Short. 1985. Major polyadenylated transcripts of cassava latent virus and location of the gene encoding coat protein. EMBO J. 4:33-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Townsend, R., J. Watts, and J. Stanley. 1986. Synthesis of viral DNA forms in Nicotiana plumbaginifolia protoplasts inoculated with cassava latent virus (CLV): evidence for the independent replication of one component of the CLV genome. Nucleic Acids Res. 14:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Wezel, R., H. Liu, P. Tien, J. Stanley, and Y. Hong. 2001. Gene C2 of the monopartite geminivirus tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localised in the nucleus. Mol. Plant-Microbe Interact. 14:1125-1128. [DOI] [PubMed] [Google Scholar]
- 52.van Wezel, R., X. Dong, P. Blake, J. Stanley, and Y. Hong. 2002. Differential roles of geminivirus Rep and AC4 (C4) in the induction of necrosis in Nicotiana benthamiana. Mol. Plant Pathol. 3:461-471. [DOI] [PubMed] [Google Scholar]
- 53.van Wezel, R., X. Dong, H. Liu, P. Tien, J. Stanley, and Y. Hong. 2002. Mutation of three cysteine residues in tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and post-transcriptional gene silencing suppression. Mol. Plant-Microbe Interact. 15:203-208. [DOI] [PubMed] [Google Scholar]
- 54.van Wezel, R., H. Liu, Z. Wu, J. Stanley, and Y. Hong. 2003. Contribution of the zinc finger to zinc and DNA binding by a suppressor of posttranscriptional gene silencing. J. Virol. 77:696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Arnim, A., and J. Stanley. 1992. Inhibition of African cassava mosaic virus systemic infection by a movement protein from the related geminivirus tomato golden mosaic virus. Virology 187:555-564. [DOI] [PubMed] [Google Scholar]
- 57.von Arnim, A., T. Frischmuth, and J. Stanley. 1993. Detection and possible functions of African cassava mosaic virus DNA B gene products. Virology 192:264-272. [DOI] [PubMed] [Google Scholar]
- 58.Zhan, X., A. Haley, K. Richardson, and B. Morris. 1991. Analysis of the potential promoter sequences of African cassava mosaic virus by transient expression of the β-glucuronidase gene. J. Gen. Virol. 72:2849-2852. [DOI] [PubMed] [Google Scholar]