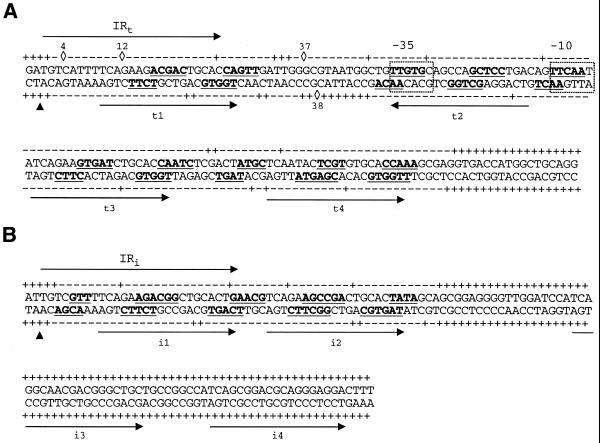
Figure 6.
Summary and details of the chemical and enzymatic probe protection analyses of the TniA contacts with the t-end (A) and the i-end (B). The arrows above the sequences indicate the terminal 25 bp inverted repeats while the arrows under the sequences indicate the 19 bp repeats. Transposase nicking sites are indicated with filled triangles. Sequences protected from hydroxyl radical cleavage are in bold and underlined. The linkages marked – were protected from DNase I cleavage. Phosphodiester linkages cleaved by DNase I are indicated with +, while DNase I hypersensitive phosphates are indicated with diamonds and the number of the nucleotide starting from the transposon termini. The data for DNase I footprinting of i3 and i4, illustrated here, are not shown in Figure 4. Promoter hexamers are boxed.