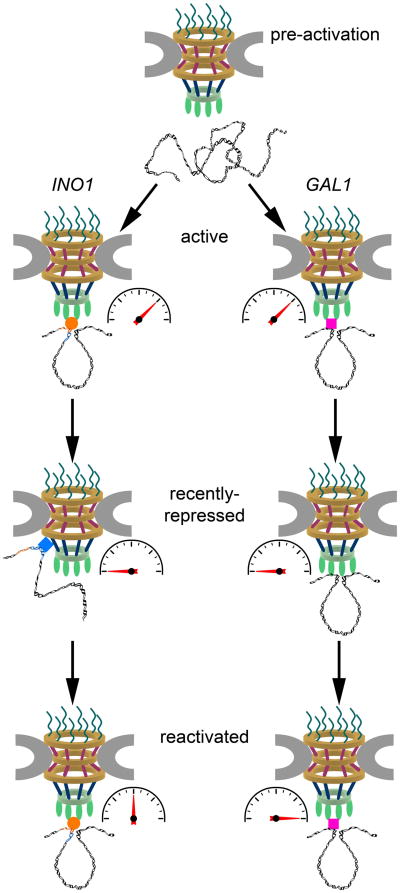
FIGURE 1. Models for Transcriptional Memory at the NPC.
Prior to transcriptional activation, the INO1 and GAL1 genes are located in the nuclear interior. Upon transcriptional activation, both genes loop and relocate to the nuclear periphery where they associate with components of the nuclear basket of the NPC, such as Nup60, Mlp1, and Mlp2. GRS I and GRS II mediate these interactions between INO1 and the NPC through an as-yet unidentified factor (orange circle). Transcriptional activators including the SAGA complex mediate interactions between GAL1 and the NPC (pink square). Following transcriptional shutoff, INO1 loses its gene loop and associates with the Nup84 subcomplex and Nup100 within the NPC core. The MRS promotes these interactions and the incorporation of H2A.Z through an unknown MRS-binding factor (blue diamond). In contrast, GAL1 maintains both its gene loop and its association with Mlp1 upon transcriptional repression. Gene induction rates in each case are indicated by gauges. Re-induction kinetics for recently repressed INO1 are slower than initial INO1 activation, whereas re-induction of recently repressed GAL1 is more rapid than initial GAL1 activation.