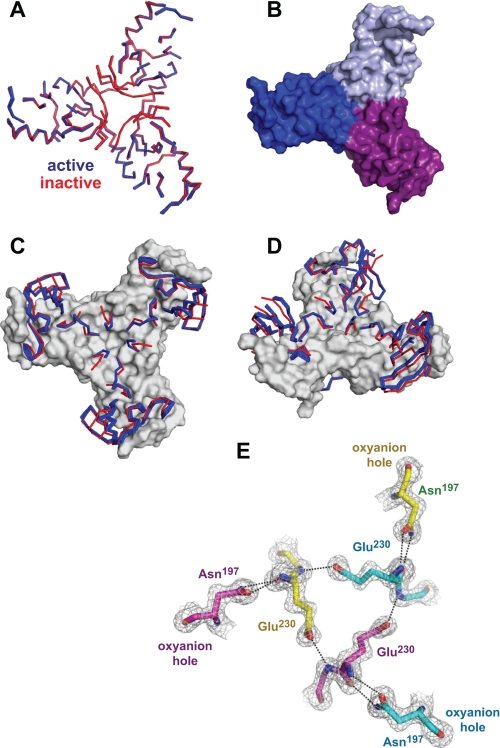
FIGURE 6.
Common and varied features in active and inactive DegSΔPDZ structures. A, ribbon representation of backbone positions in the “base” (residues 45–58, 82–84, 90–93, 117–121, 139–146, 151–159, 167–175, 189–194, 204–216, and 233–249) that are highly conserved following structural alignment of seven active (blue) and five inactive (red) DegSΔPDZ trimers. B, surface representation of the trimer base from the wild-type structure (Protein Data Bank code 3LGI). C and D, ordered backbone positions that switch conformation between the active (blue) and inactive (red) structures are shown in ribbon representation. The conserved base is shown in surface representation. E, model and electron density (1.5 σ) for residues Asn197, Glu230, and Gly231 in the wild-type DegSΔPDZ trimer (Protein Data Bank code 3LGI): yellow carbons, chain A; cyan carbons, chain B; purple carbons, chain C. All active trimers contain the same network of hydrogen bonds (dashed lines), which link the oxyanion holes of different subunits.