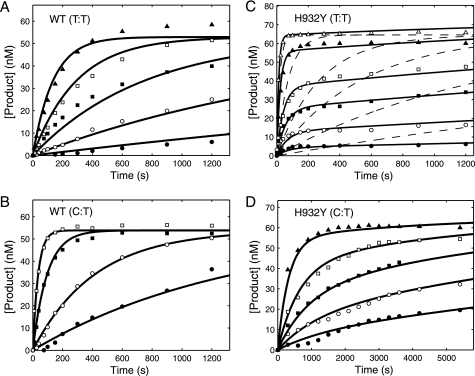
FIGURE 3.
Kinetics of misincorporation by WT enzyme and H932Y mutant. For each concentration dependence, a preformed enzyme-DNA complex ([enzyme] > [DNA duplex]) was rapidly mixed with Mg2+ and various concentrations of incorrect nucleotide. The time course of product formation was then fit globally. In each experiment, the final concentrations of the enzyme and DNA after mixing were 100–150 and 75 nm, respectively. In globally fitting each data set, the concentration of active enzyme was adjusted to fit the amplitude. A, formation of a T:T mismatch by WT exo− pol γ at each TTP concentration (1.5 (●), 5 (○), 15 (■), 50 (□), and 250 (▴) μm) was globally fit to the mechanism shown in Scheme 1, yielding an apparent Kd of 81.8 ± 10.9 μm and kpol of 0.01 ± 0.004 s−1. B, formation of a C:T mismatch by WT exo− pol γ at each dCTP concentration (15 (●), 50 (○), 250 (■), and 1000 (□) μm) was fit globally to the mechanism shown in Scheme 1, yielding an apparent Kd of 1030 ± 193 μm and kpol of 0.06 ± 0.018 s−1. C, formation of a T:T mismatch by H932Y exo− mutant at each TTP concentration (15 (●), 50 (○), 125 (■), 250 (□), 1000 (▴), and 5000 (△) μm) was fit globally (solid line) to the mechanism shown in Scheme 2, yielding an apparent Kd of 1630 ± 310 μm, k2 of 0.1 ± 0.02 s−1, k−2 of 0.01 ± 0.003 s−1, and k3 of 0.0004 ± 0.0005 s−1. The dashed line indicates an attempt to fit the data with the mechanism shown in Scheme 1, showing that it cannot account for the concentration dependence of the rate and amplitude. D, formation of a C:T mismatch by H932Y exo− mutant at various dCTP concentrations (175 (●), 375 (○), 750 (■), 1500 (□), and 4000 (▴) μm) were globally fit to the mechanism shown in Scheme 2, yielding an apparent Kd of 22200 ± 3420 μm, k2 of 0.02 ± 0.005 s−1, k−2 of 0.0004 ± 0.003 s−1, and k3 of 0.0003 ± 0.002 s−1. Traces for the formation of a G:T mismatch are not shown, but all time courses were fit to the mechanism shown in Scheme 1, and the resulting parameters are summarized in Table 2.