Abstract
In most mammalian cells, the retinoic acid receptor (RAR) is nuclear rather than cytoplasmic, regardless of its cognate ligand, retinoic acid (RA). In testis Sertoli cells, however, RAR is retained in the cytoplasm and moves to the nucleus only when RA is supplied. This led us to identify a protein that regulates the translocation of RAR. From yeast two-hybrid screening, we identified a novel RAR-interacting protein called CART1 (cytoplasmic adaptor for RAR and TR). Systematic interaction assays using deletion mutants showed that the C-terminal CoRNR box of CART1 was responsible for the interaction with the NCoR binding region of RAR and TR. Such interaction was impaired in the presence of ligand RA, as further determined by GST pulldown assays in vitro and immunoprecipitation assays in vivo. Fluorescence microscopy showed that unliganded RAR was captured by CART1 in the cytoplasm, whereas liganded RAR was liberated and moved to the nucleus. Overexpression of CART1 blocked the transcriptional repressing activity of unliganded apoRAR, mediated by corepressor NCoR in the nucleus. CART1 siRNA treatment in a mouse Sertoli cell line, TM4, allowed RAR to move to the nucleus and blocked the derepressing function of CART1, suggesting that CART1 might be a cytoplasmic, testis-specific derepressor of RAR.
Keywords: Gene Regulation, Hormone Receptors, Protein Translocation, Protein-Protein Interactions, Repressor Protein, Cytoplasmic Retention, NCoR, RAR, Sertoli Cells, Derepressor
Introduction
Nuclear receptors (NRs)3 are ligand-dependent transcription factors that control diverse aspects of development and homeostasis by regulating expression of their target genes (1, 2). The NR superfamily, which shares a common structural organization composed of distinct domains, is classified into three groups, steroid hormone receptors, non-steroid hormone receptors, and orphan receptors. One of the major regulatory effects of NRs can be achieved by a change in subcellular location in response to various cellular factors. Steroid hormone receptors, such as glucocorticoid receptor and estrogen receptor, reside in the cytoplasm in association with heat shock protein 90 (Hsp90) but on the entry of their cognate ligands dissociate from Hsp90 and translocate to the nucleus to exert transcriptional activation (3–6). However, non-steroid hormone receptors, such as RAR and TR, reside solely in the nucleus regardless of their ligands in most mammalian cells. In the absence of ligands, these receptors transverse the nuclear membrane and associate with nuclear corepressors, such as nuclear receptor corepressor (NCoR1) or silencing mediator for retinoid and thyroid hormone receptors (SMRT or NCoR2) to mediate transcriptional repression (7, 8). These corepressors then recruit histone deacetylase, resulting in histone deacetylaton, chromatin compaction, and silencing of target gene expression (9, 10). In the presence of ligands, corepressors dissociate, and instead, coactivators with histone acetyltransferase activity associate to mediate transcriptional activation (11, 12).
Despite significant progress in understanding nuclear regulation of RAR and TR, little is known about their regulation in the cytoplasm. A few reports have suggested that TRs (TRα1 and TRβ1) actively shuttle between the nucleus and cytoplasm and that the ligand promotes cytoplasm-to-nucleus shuttling (13, 14). In some Sertoli cell lines derived from mouse testis, RARα is ordinarily located in the cytoplasm and rapidly moves to the nucleus in response to its ligand RA (15, 16). In both cases, ligands are required for the promotion of nuclear translocation. However, it is largely unknown what cellular factor(s) is responsible for the cytoplasmic retention of TRs and RARα in the absence of the ligands.
Recently, Rab11-FIP3 (also known as arfophilin and eferin) was identified by a yeast two-hybrid screen as an effector protein of Rab11 and ADP-ribosylation factor ARF5/6, both of which are members of the small GTPase family that functions in endosomal recycling and intracellular membrane trafficking (17–19). In particular, Rab11 plays an essential role in protein recycling from endosomes to the plasma membrane (20). Similarly, ARFs are key regulators of membrane trafficking and the actin cytoskeleton. FIP3 simultaneously interacts with Rab11 and ARF GTPases. This interaction is mediated by a highly conserved Rab11-binding domain (RBD) located at the C-terminal end of FIP3 (18, 21, 22), which is also important for ARF5 binding (23). Recently, structural studies revealed that two Rab11 molecules bind to dyad symmetrical sites at the C terminus of a FIP3 dimer (21, 22). Additionally, the amphiphilic α-helix of FIP3-Rab11 binding domain at the C terminus raises the possibility that FIP3 may accommodate binding partners other than Rab11 and ARF5/6.
In this study we identified Rab11-FIP3 as a novel RAR binding partner using a yeast two-hybrid system. Subsequently, we demonstrated that Rab11-FIP3 interacted with RAR and TR among the NRs tested through its conserved C terminus. As noted above, Rab11-FIP3 has also been called arfophilin and eferin. Hereinafter, we refer to it as “cytoplasmic adaptor of RAR and TR” (CART1). Extensive in vitro and in vivo assays revealed that CART1 associates with unliganded RAR (and TR) and dissociates from RAR in the presence of ligand. CART1 binding to RAR (and TR) is similar to NCoR binding, but it is different in that CART1 is cytoplasmic, whereas NCoR is nuclear. The RAR binding motif of both CART1 and NCoR is highly conserved, as shown by binding assays with CART1 mutants and competition assays. CART1 overexpression blocks RAR entry into the nucleus and, thus, impairs the transcriptional repressing activity of unliganded RAR, likely mediated by NCoR. In contrast, CART1 knockdown in the mouse Sertoli cell line TM4 resulted in significant augmentation of the repressing potential of RAR. Our findings suggest that CART1 may be a cytoplasmic, testis-specific derepressor of RAR, which could be an additional checkpoint in the fine control of gene expression.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
NIH3T3 and TM4 cell were maintained in DMEM medium supplemented with 5% heat-inactivated fetal bovine serum (FBS) and antibiotic-antimycotic mix (all from Invitrogen). For transcription assays, FBS was pretreated with charcoal.
Plasmids and Cloning
All cDNAs were made according to standard methods and verified by sequencing. The multicopy yeast expression plasmids used in the two-hybrid assays were pBTM116 and pASV3 (24). The human CART1 full-length cDNA was kindly provided as a KIAA clone (KIAA0665) by the Kazusa DNA Research Institute (Japan). Deletion and point mutants of CART1 were created by PCR amplification and subcloned into the pBTM116 or pASV3 vectors. FLAG (2×)-tagged cTRβ, mRARβ, and CART1 genes were inserted in the pcDNA3 vector (Invitrogen). Green fluorescence protein (GFP)- and red fluorescence protein (HcRed)-tagged constructs were inserted into pEGFP-C3 and pHcRed (BD Biosciences), respectively. For GST-fused CART1, pGEX2T (Amersham Biosciences) was used.
Yeast Two-hybrid Screening and Assays
A human HeLa cDNA library in the prey plasmid pACT2 (BD Biosciences) was screened for proteins that interact with the ligand binding domain (DEF region) of hRARα using the yeast reporter strain L40. The experimental procedures were the same as those previously described (25), except that the ligand all-trans retinoic acid (AtRA) was omitted. To map the interaction domains in CART1, deletion derivatives of CART1 were fused with the VP16 AD by subcloning into pASV3. To determine the specificity of the interaction, other NR family members, including RXRα, estrogen receptor α, glucocorticoid receptor, and TRβ, were subcloned into pBTM116. The resulting VP16 AD-CART1 fusion vectors were co-introduced with the pBTM116 derivatives encoding the LexA DBD-NR fusion proteins into L40 cells. The level of interaction was determined by quantitative β-galactosidase assays.
Glutathione S-Transferase Pulldown Assays
A GST fusion of CART1 (amino acids 699–756) was purified on glutathione-Sepharose beads (Amersham Biosciences) by standard methods. Indicated NR proteins were in vitro translated in 50 μl of rabbit reticulocyte lysate (Promega, Madison, WI) supplemented with [35S]methionine (Amersham Biosciences). For competition assays, a fragment of NCoR1 (amino acids 1953–2440) was synthesized. Other experimental procedures have been described previously (26, 27). The bound proteins were eluted by boiling in sample buffer for 5 min and visualized by SDS-PAGE followed by autoradiography.
Immunoprecipitation (IP) Analysis
After transfection with the indicated plasmid DNA using the Lipofectamine plus reagent (Invitrogen), NIH3T3 cells were washed in phosphate-buffered saline (PBS), and cell lysates were prepared by adding 1 ml of TEN-modified buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 0.1% Nonidet P-40, 5 mm EDTA, 1 mm PMSF) supplemented with protease inhibitors (Roche Molecular Biochemicals). Lysates were precleared by preincubation with protein A/G-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) for 15 min and incubated at 4 °C for 2 h with beads and a 1:200 dilution of anti-RARα antibody (all from Santa Cruz Biotechnology). Beads were then washed once with TEN and twice with PBS, and the immune complexes were released from the beads by boiling in sample buffer for 5 min. After electrophoresis on 10% SDS-PAGE, immunoprecipitated products were analyzed by Western blotting (WB) using anti-FLAG M2 monoclonal antibody (F-3165; Sigma; 1:1000). The endogenous interaction between RARα and CART1 in TM4 cells was determined by IP with anti-CART1 antibody (rabbit polyclonal serum against amino acids 206–719 of human CART1) and subsequent WB with anti-RARα antibody. For control, IgG (Santa Cruz Biotechnology) was used.
Fluorescence Microscopy
NIH3T3 cells seeded on coverslips were transfected with pEGFP-C3 for GFP-CART1 and pHcRed-C1 for HcRed-mRARβ (or cTRβ). One day later cells were treated with 1 μm AtRA (or T3) and incubated for 24 h. Cells were then washed with PBS and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After treatment with 10 μl of VectaShield (Vector Laboratories, Burlingame, CA), cells were observed under a fluorescence microscope (Olympus Optical Co.). DAPI (1 μg/ml; Sigma) was used to localize chromosomal DNA in the nucleus. To locate endogenous RARα, either NIH3T3 or TM4 cells was fixed with paraformaldehyde and permeabilized for 10 min at 4 °C in PBS containing 0.1% Triton X-100. After washing, cells were blocked for 60 min in PBS containing 2% bovine serum albumin and then incubated for 2 h with rabbit anti-RARα polyclonal antibody (1:100; Santa Cruz Biotechnology) diluted in blocking buffer. Cells were then incubated with Texas Red-conjugated anti-rabbit antibody (1:200; Santa Cruz Biotechnology) for 1 h in the dark, mounted with VectaShield, and observed by fluorescence microscopy.
Transient Transfection and Luciferase Reporter Assays
NIH3T3 cells were seeded in a 12-well culture plate. Transient transfections were performed using the Lipofectamine plus reagent (Invitrogen) with an RARα expression vector, RARE-tk-Luciferase reporter, and SV40-driven β-galactosidase expression vector as an internal control. Then, 4 h after transfection, cells were washed, fed with 5% charcoal-striped medium, and incubated for an additional 16 h. Depending on the experimental conditions, 1 μm AtRA was added. Cells were then washed with ice-cold PBS, collected, resuspended in 50 μl of luciferase lysis buffer (Promega), and subjected to three freeze-thaw cycles. Luciferase activity was measured by adding 20 μl of luciferin into 30 μl of cell lysate and using an analytical luminescence luminometer according to the manufacturer's instructions (Promega). The β-galactosidase activity was determined in 96-well plates using a microplate reader at 405 nm. The luciferase activity was normalized to the β-galactosidase activity.
Reverse Transcriptase and Real-time PCR
TM4 cells were transfected with control siRNA or CART1-specific siRNA for knockdown. Total RNA was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. RNA (1.5 μg) was reverse-transcribed into cDNA using oligo(dT)6 primers (New England Biolabs, Beverly, MA) and Moloney murine leukemia virus reverse transcriptase (RT; Invitrogen). RT products were amplified by PCR using the following primer pairs: for RARβ2, forward, 5′-TTGTGTTCACCTTTGCCAAC-3′, and reverse, 5′-CGGTTCCTCAAGGTCCTGG-3′; for GAPDH, forward, 5′-CGGCTACCACATCCAAGGAA-3′, and reverse, 5′-AGCCACATCGCTCAGACACC-3′. The expression levels were normalized using GAPDH as an internal standard in each well. Relative expression (%) was defined as the variation relative to control.
RNA Interference
The sequences of the custom siRNA duplex (Stealth system, Invitrogen) for mouse CART1 were as follows: sense, 5′-UUAUUCUCCUGUUCCUCACUGAGCC-3′, and antisense, 5′-GGCUCAGUGAGGAACAGGAGAAUAA-3′. The siRNA duplex control used was the Stealth RNAi Negative Control with medium GC content (Invitrogen). Transfection of the siRNAs into TM4 cells was performed with Lipofectamine 2000 in Opti-MEM I reduced-serum medium (Invitrogen) according to the manufacturer's instructions. In independent experiments, transfection efficiency was assessed to be greater than 80% using fluorescein-labeled siRNA (Invitrogen). Knockdown of mouse CART1 was verified by WB using an anti-CART1 antibody.
RESULTS
Identification of CART1
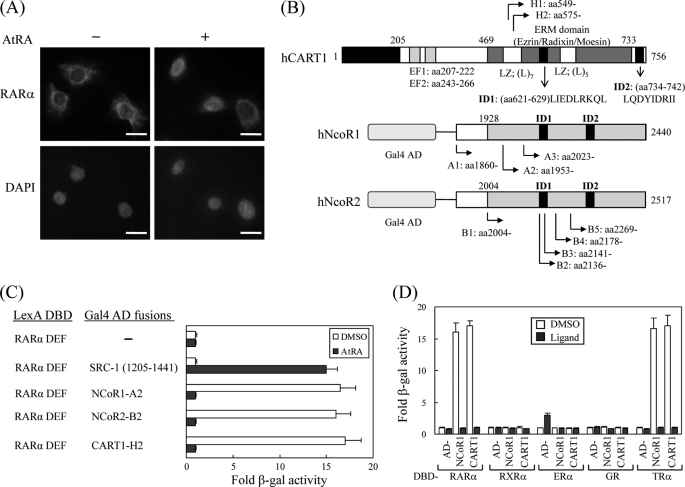
In most mammalian cells, RAR and TR are nuclear rather than cytoplasmic regardless of their cognate ligands. However, some studies have demonstrated that RAR and TR are retained in the cytoplasm and move to the nucleus when ligands are supplied (13–16). In particular, we also found that RARα shuttles between the cytoplasm and the nucleus in the Sertoli cell line TM4 in response to AtRA (Fig. 1A). However, it is largely unknown what cellular factor(s) is responsible for the cytoplasmic retention of TRs and RARα. To identify such a factor(s), we performed a yeast two-hybrid screen of a HeLa cDNA library using the ligand binding domain (or DEF) of hRARα as bait. From the screen, we isolated 101 clones that were His gene-positive and showed high β-galactosidase activity. Plasmids from these clones were recovered and sequenced. GenBankTM search results indicated 8 NCoR1, 13 NCoR2, 63 CART1, and 17 RXR with overlapping sequences (Fig. 1B). Of these, RXR is a heterodimer partner of RAR, and NCoRs have been reported as corepressors of RAR and TR (7, 8), indicating that our screen was reliable. Most of the clones were found to be CART1 (also known as Rab11-FIP3, arfophilin, and eferin), which is expected to have a role in protein recycling and the actin cytoskeleton (17–19). Like the NCoR-RARα interaction, CART1 strongly interacts with RARα, and the interaction is abolished in the presence of AtRA (Fig. 1C). Subsequent yeast two-hybrid assays indicated that CART1 interacted with TRβ among the NR members tested (Fig. 1D). CART1 also interacted with other isoforms of RARα and TRβ with comparable binding affinities (data not shown). This property of CART1 is similar to that of NCoR, suggesting that CART1 may contain a conserved CoRNR box that has been identified as a RAR/TR binding motif in NCoRs (28–30). In fact, we found two putative CoRNR boxes at amino acid residues 621–629 and 734–742 (Fig. 1B).
FIGURE 1.
Isolation of CART1. A, shown is subcellular localization of RARα in TM4 cells. Mouse RARα in TM4 cells were visualized by staining with rabbit anti-RARα polyclonal antibody and Texas Red-conjugated anti-rabbit antibody in the absence and presence of ligand AtRA. Each bar represents 10 μm. B, shown are schematic representations of human CART1, NCoR1, and NCoR2 (SMRTe) isolated by yeast two-hybrid screening. Locations of identified clones and functional domains are indicated: EF, EF-hand motif; LZ, leucine zipper; ID, nuclear receptor interaction domain. C, interaction of CART1 with hRAR(DEF) is shown. Interactions were monitored by introducing LexA DBD-fused RAR(DEF) and Gal4 AD-fused test partners in yeast strain L40 and by β-galactosidase (β-gal) assays with transformed yeast extracts. -Fold β-galactosidase activity indicates relative value compared with the Gal4 AD empty control. In all experiments, 1 μm AtRA was added to the yeast culture. DMSO was used as a control for AtRA. Data are shown as the averages of three independent experiments (mean ± S.D.). D, shown is interaction of CART1 with other nuclear receptor family members. Yeast two-hybrid assays were performed with the other NRs indicated in the presence of their cognate ligands: RAR, 1 μm AtRA; RXR, 1 μm 9-cis RA; ER, 1 μm estradiol (E2); GR, 5 μm deoxycorticosterone; TR, 1 μm triiodo-l-thyronine (T3). Data are the average of three independent experiments (mean ± S.D.).
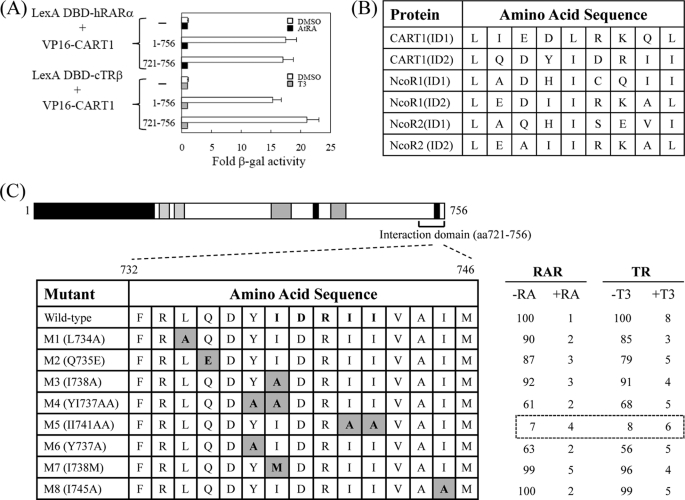
Mapping the Interaction Domain between CART1 and RAR/TR
To map the RAR/TR-interacting domain in CART1, genes for its deletion derivatives were placed in the pASV3 vector. Subsequent yeast two-hybrid assays indicated that the region bounded by amino acids 721–756 was the minimal domain that was necessary and sufficient for binding to RARα and TRβ (Fig. 2A). As expected, the interaction was diminished in the presence of ligands AtRA for RARα and T3 for TRβ. The amino acid sequences of two putative CoRNR boxes of CART1 were compared with those of NCoR1 and NCoR2, where the consensus sequence corresponding to the CoRNR box is (L/I)XX(I/V)IXXXL (Fig. 2B). The amino acid alignment indicated that the C-terminal CoRNR box of CART1 was well conserved in mammals (data not shown). To determine which amino acid residues in that region are critical for the interaction with RAR and TR, eight point mutants were generated by PCR and subcloned into pASV3. As shown in Fig. 2C, one mutant (M5) with AA substitutions from two central isoleucines was defective in RAR/TR binding. These isoleucines are well conserved in the CoRNR boxes of NCoR1 and NCoR2, suggesting that CART1 and NCoRs share a common binding motif for RAR and TR binding.
FIGURE 2.
Mapping of RAR binding domain in CART1. To map the minimal region required for RAR and TR binding (A), CART1 deletions were fused to acidic VP16 AD using the pASV3 vector instead of Gal4 AD. Using these CART1 deletions and LexA DBD-RAR(DEF) or LexA DBD-TR(DE), yeast two-hybrid assays were performed in the absence (DMSO) and presence of AtRA (for RAR) or T3 (for TR). -Fold β-galactosidase activity indicates relative values compared with the VP16 AD empty control. B, shown is amino acid alignment of CART1, NCoR1, and NCoR2. Amino acid sequences of interacting domain (ID or CoRNR box) are listed. The consensus sequence corresponding to CoRNR box is (L/I)XX(I/V)IXXXL. C, shown is identification of critical amino acid residues of CART1 responsible for RAR or TR binding. The amino acid residues (aa) in the second putative CoRNR box (also designated as ID2) were mutated to alanine by PCR. Yeast two-hybrid assays were performed using CART1 mutants and LexA DBD-RAR(DEF) or LexA DBD-TR(DE). Numbers represent the relative percent of wild-type β-galactosidase activity shown by the average of three independent experiments (mean ± S.D.).
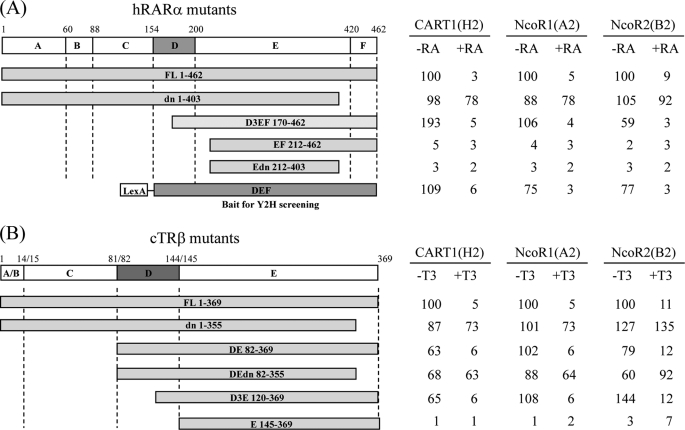
To map the CART1-interacting domain in RARα and TRβ, deletion mutants of RARα and TRβ were fused to the LexA DBD using the pBTM116 vector and assayed for interaction with Gal4 AD-CART1 (amino acids 575–756) in yeast L40. For comparison, Gal4 AD-NCoR1 (1953–2440) and Gal4 AD-NCoR2 (2141–2517) were used. The interaction of NCoR with RAR and TR has been shown to require a conserved sequence lying within the α-helix H1 of the ligand binding domain (DEF or DE) (7, 31, 32). Like NCoR1 and NCoR2, CART1 failed to interact with RARα and TRβ when helix H1, corresponding to the D3 region in Fig. 3, was removed (compare DEF and D3EF with EF for RAR and DE and D3E with E for TR; Fig. 3). The C-terminal deletion of helix H12 containing AF-2 AD core, as shown by the dominant negative, had little effect on the constitutive interaction with CART1 but abolished the ligand-dependent decrease of the CART1-RARα and CART1-TRβ interactions. CART1 also interacted with v-erbα, a viral version of TR defective in AF-2 AD without ligand effect (data not shown). A similar pattern of the interaction was observed for NCoR1 and NCoR2. Thus, these results suggest that helix H1 (or the D3 region) of RAR or TR is required for CART1 interaction, whereas helix H12 (or AF-2 AD) is dispensable for the interaction but essential for ligand-dependent disruption of the interaction. Combined, our mapping data indicate that the CART1 interaction mode with apo (unliganded) and holo (liganded) RARα and TRβ is reminiscent of the NCoR mode, with the same or very similar binding surfaces.
FIGURE 3.
Comparison of the interaction modes of CART1, NCoR1, and NCoR2. The interactions were monitored by yeast two-hybrid assays using Gal4 AD-CART1 (clone H2), -NCoR1 (clone A2), or -NCoR2 (clone B2) and LexA DBD-RARα mutants (A) or LexA DBD-TRβ mutants (B). The range of amino acids in each mutant is indicated. Functional domains of RARα and TRβ were designated as A–F (or E for TR). Numbers represent the relative percent of wild-type β-galactosidase activity.
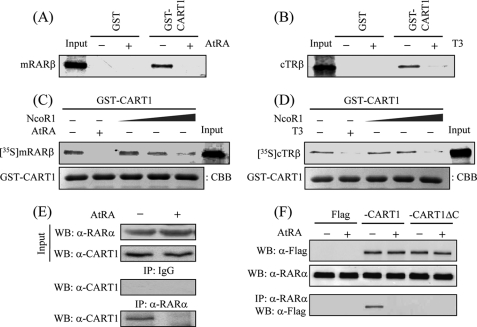
Confirmation of the Interaction by GST Pulldown and IP Assays
For GST pulldown assays, GST-fused CART1 (amino acids 699–756: containing the second CoRNR box) was expressed in Escherichia coli, purified, and mixed separately with in vitro translated, 35S-labeled mRARβ or cTRβ. Consistent with the yeast two-hybrid data, the 35S-labeled mRAR or cTR was retained by CART1 in the absence of the cognate ligand, and the retention was abrogated in the presence of ligand (Fig. 4, A and B). To verify the functional relevance of CART1 and NCoR binding to RAR and TR, competition assays were performed using GST-CART1, synthesized 35S-cTRβ or cTRβ, and unlabeled NCoR1 (amino acids 1953–2440). As the amounts of NCoR1 increased, CART1 binding to apoRAR or apoTR decreased (Fig. 4, C and D), suggesting that NCoR1 competes with CART1 for RAR or TR binding, likely due to close similarity in binding surfaces between NCoR1 and CART1.
FIGURE 4.
Confirmation of the interaction by GST pulldown and IP assays. A and B, shown is the interaction between CART1 and RAR or TR in vitro. In vitro synthesized 35S-hRARβ (A) or 35S-cTRβ (B) was incubated with 2 μg of GST-fused hCART1 (amino acids 699–756) in the presence of cognate ligand (2 μm). Bound proteins were visualized by SDS-PAGE and autoradiography. Input was 10% of the labeled sample used in assay. C and D, competition between CART1 and NCoR1 for RAR or TR binding is shown. For assays, 10 μl of in vitro translated 35S-hRARβ (C) or 35S-cTRβ (D) was mixed with purified GST-CART1 (amino acids 699–756) and further reacted with increasing volumes of NCoR1 (10, 20, and 40 μl). Then, GST pulldown assays were performed as described. Equal loading of beads was shown by Coomassie Brilliant Blue (CBB) staining of GST-CART1. E, endogenous interaction between CART1 and RARα is shown. TM4 cell lysates were prepared and immunoprecipitated with preimmune serum (IgG) or anti-RARα antibody. Precipitated proteins were revealed by WB using anti-CART1 antibody. F, requirement of the C-terminal region of CART1 for RAR binding in vivo is shown. NIH3T3 cells were transfected with FLAG-hCART1 or CART1 C-terminal truncation (CART1ΔC) expression vector and cultured in the absence and presence of 1 μm AtRA. The interaction was monitored by IP with anti-RARα antibody and WB using anti-FLAG antibody.
The endogenous interaction between RARα and CART1 in TM4 cells was demonstrated by IP with anti-CART1 antibody and subsequent WB using anti-RARα antibody (Fig. 4E). Other co-IP assays in NIH3T3 cells using exogenously expressed FLAG-tagged human full-length CART1 or the C-terminal deletion (CART1ΔC) further emphasized that the C-terminal region of CART1 is essential for RARα interaction in vivo (Fig. 4F). In both IPs, the interactions were disrupted in the presence of AtRA. These in vitro and in vivo findings confirmed the interaction observed in yeast, further emphasizing that both CART1 and NCoR share a common motif for binding to apoRAR (and apoTR).
Effect of CART1 on Cytoplasmic Localization of RAR
Our data showed that CART1 associated with apoRAR or apoTR and dissociated from the complex in the presence of ligand. Next, we examined the effect of the interaction on the subcellular localization of RAR. Previous studies have indicated that RAR is predominantly nuclear in most mammalian cells (33–35), but CART1 is a cytoplasmic protein that locates in the pericentrosomal region (17–19). To determine the effect of CART1 on the subcellular localization of RAR in living cells, we appended the GFP to human CART1 and the HcRed to mouse RARβ for fluorescence microscopy. When HcRed-RAR alone was expressed in NIH3T3 cells, RAR was nuclear (Fig. 5A). However, when both were transfected, RAR moved to the cytoplasm together with CART1 (Fig. 5B). Upon AtRA treatment, RAR was released from CART1, and RAR translocated to the nucleus, whereas CART1 remained in the cytoplasm. Similar subcellular distribution and ligand effects were observed when HcRed-TRβ was used (data not shown).
FIGURE 5.
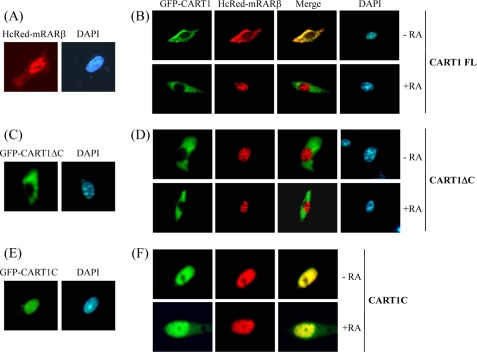
Cytoplasmic retention of RAR by CART1. NIH3T3 cells were transfected with HcRed-mRARβ and GFP-CART1 (or its variants) alone or together and treated with 1 μm AtRA. Cells were then fixed and observed under a fluorescence microscope. DAPI (1 μg/ml) was used to localize chromosomal DNA in the nucleus. A and B, localization of HcRed-RARβ alone (A) and together with GFP-CART1 (B) is shown. C and D, localization of CART1 C-terminal truncation (CART1 ΔC: amino acids 1–719) alone (C) and together with RAR (D) is shown. E and F, localization of the C-terminal region of CART1 (CART1C: amino acids 699–756) alone (E) and together with RAR (F) is shown.
To determine whether the CART1-RAR interaction was essential for the cytoplasmic retention of RAR, the CART1 C-terminal truncation (CART1ΔC: amino acids 1–719), defective in RAR binding, was fused to GFP and expressed alone or coexpressed with HcRed-RARβ in NIH3T3 cells. CART1ΔC alone was cytoplasmic (Fig. 5C). When both were expressed, RAR exhibited constitutive nuclear localization regardless of AtRA, leaving CART1 in the cytoplasm (Fig. 5D). This result supports the notion that CART1 binding is required to keep RAR in the cytoplasm. In contrast, the C-terminal region of CART1 (CART1C: amino acids 699–756) was constitutively nuclear regardless of expression alone (Fig. 5E) or together with RAR (Fig. 5F), suggesting that RAR associates with CART1C and moves to the nucleus together. Thus, the other region of CART1 may be responsible for the cytoplasmic retention of CART1. Overall, our data demonstrate that CART1 anchors RAR (or TR) in the cytoplasm through a protein-protein interaction and liberates RAR to the nucleus through dissociation in the presence of ligand.
Cytoplasmic Distribution of Endogenous RAR in Testis Sertoli Cells
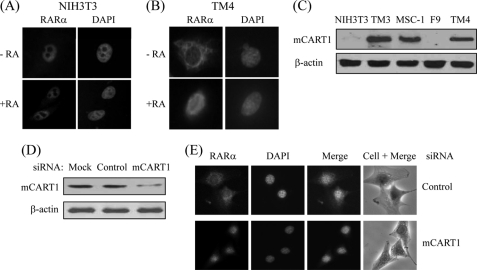
It has been reported that apoRARα is cytoplasmic and holo-RARα is nuclear in mouse testis Sertoli cell lines (15, 16). These findings led us to investigate the role of CART1 in the intracellular distribution of RARα in Sertoli cells. For that purpose, we first compared the locations of endogenous RARα in mouse NIH3T3 cells and the Sertoli cell line TM4. As shown in Fig. 6, A and B, RARα was constitutively nuclear regardless of its ligand AtRA in NIH3T3 cells, whereas RARα shuttled from cytoplasm to nucleus in response to AtRA in TM4 cells. To determine whether CART1 was responsible for this difference in subcellular location of RARα between two cells, the expression of CART1 was measured by WB. CART1 was predominantly expressed in three testis cell lines (Leydig cell, TM3; Sertoli cell, MSC-1 and TM4) of five mouse cell lines tested (Fig. 6C). Thus, to see the effect of CART1 knockdown on the subcellular distribution of RARα, we chose TM4 cells that express CART1 in the cytoplasm (supplemental Fig. 1) and treated with siRNA (determined by WB; Fig. 6D). As shown in Fig. 6E, CART1 knockdown led to relocation of RARα to the nucleus even in the absence of AtRA. A similar knockdown effect of CART1 on RAR localization was observed in other Sertoli cell MSC-1 (supplemental Fig. 2). Taken together, these results suggest that CART1 is an important mediator for the cytoplasmic retention of apoRARα in Sertoli cells.
FIGURE 6.
Effect of CART1 knockdown on subcellular location of RAR in mouse Sertoli cells. A and B, subcellular location of endogenous RARα in NIH3T3 and TM4 Sertoli cells is shown. NIH3T3 (A) or TM4 (B) cells were fixed and permeabilized. Cells were then stained with rabbit anti-RARα polyclonal antibody and Texas Red-conjugated anti-rabbit antibody and observed by fluorescence microscopy. C, expression of CART1 is shown. Cellular extracts were prepared from mouse cell lines as indicated and subjected to WB using anti-CART1 antibody. β-Actin was used as an internal control. D and E, effect of CART1 knockdown on subcellular location of RAR in TM4 cells. TM4 cells were transfected with either control or CART1 siRNA (200 pmol) using Lipofectamine 2000 in the absence of AtRA. CART1 expression was monitored by WB (D). Endogenous RARα was visualized as described in B (E).
Effect of CART1 on Transrepression Activity of RAR
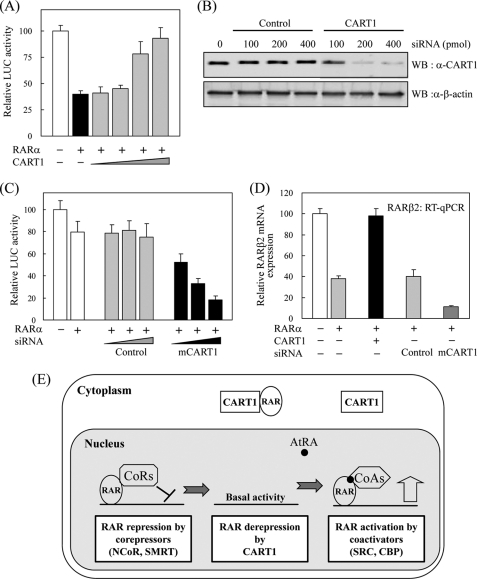
In the absence of ligand, RAR and TR can suppress or silence the basal transcriptional activity of target genes (36–38). This transrepression is due to ligand-independent association of their ligand binding domain with NCoRs in the nucleus (7, 8). First, the repressor activity of RAR and the effect of CART1 on RAR activity were monitored by transfection assays using RARE-tk-luciferase reporter and RARα in CART1-deficient NIH3T3 cells. Compared with control, RARα transfection showed strong repression in luciferase activity. When CART1 expression vector was added, the transrepressing activity of RARα was significantly impaired in a dose-dependent manner (Fig. 7A). However, no obvious effect of CART1 was observed on the AtRA-induced RARα activity (data not shown). Next, reverse assays were conducted using CART1 siRNA in CART1-abundant TM4 cells. As monitored by WB, the expression of CART1 was greatly reduced by siRNA treatment (Fig. 7B). Different from NIH3T3 cells, the transrepression activity of RARα was less severe in TM4 cells. Upon CART1 knockdown, the repressing activity of RARα became more severe than that of untreated or control condition (Fig. 7C). Finally, we assessed the effect of CART1 overexpression or knockdown on the expression of the endogenous RAR target gene RARβ2 by real-time RT-PCR. CART1 overexpression mostly recovered RAR-mediated repression of RARβ2 repression, whereas CART1 knockdown increased RARβ2 repression about 3-fold compared with control siRNA treatment (Fig. 7D). These results suggest that reduction of the CART1 level in TM4 cells may cause preferred translocation of RAR to the nucleus with further association with NCoRs and, thus, more transcriptional repression (Fig. 7E). The NCoR1 expression in TM4 cells supports this possibility (supplemental Fig. 3).
FIGURE 7.
Effect of CART1 on transcriptional activity of RAR. A, shown is the effect of CART1 on transcriptional activity of RAR in the absence of AtRA. NIH3T3 cells were cotransfected with RARα (0.2 μg) and increasing amounts (0.1, 0.2, 0.4, 0.8 μg) of FLAG-CART1 together with the RARE-tk-luciferase reporter. Each bar represents the percent of mock treatment. The relative luciferase (Luc) activity was determined by luciferase assay after normalizing to the observed β-galactosidase activity. Data are the average of three independent experiments (mean ± S.D.). B, shown is the effect of siRNA on CART1 expression. After transfection into TM4 cells, using the amounts indicated, the expression of mouse CART1 was monitored by WB using an anti-CART1 polyclonal antibody. C, shown is the effect of CART1 knockdown on transcriptional activity of RAR. TM4 cells were cotransfected with RARα (0.2 μg) and increasing amounts of CART1 siRNA, as indicated in Fig. 7C, together with the RARE-tk-luciferase reporter. The relative luciferase activity is shown by the average of three independent experiments (mean ± S.D.). D, shown is the effect of CART1 overexpression or knockdown on the expression of endogenous RAR-regulated RARβ2 gene. Total RNA was extracted from TM4 cells transfected with control or CART1-specific siRNA (200 pmol) and subjected to real-time PCR coupled to reverse transcription. The expression levels were normalized using GAPDH as an internal standard. Relative expression (%) was defined as the variation relative to control. E, shown is a postulated model for the role of CART1 in RAR regulation. In testis cells, where CART1 is abundant, CART1 interacts with apoRAR in the cytoplasm, thus preventing its repressor function by cooperating with corepressors in the nucleus. In the presence of ligand AtRA, RAR dissociates from CART1 and moves to nucleus where it interacts with coactivators for transcriptional activation. qPCR, quantitative PCR.
DISCUSSION
Understanding regulation of the subcellular localization of NRs is important in understanding NRs-mediated transcriptional control in the nucleus. It is well documented that among the NR superfamily, steroid hormone receptors are located predominantly in the cytoplasm, in association with cytoplasmic factors such as Hsp90 and kinases in the unliganded state, and translocate to the nucleus by dissociating from such factors in the presence of ligand (3–6).
In the case of non-steroid hormone receptors, such as RAR and TR, although significant progress in understanding nuclear regulation has been made (for review see Refs. 39 and 40), little is known about their regulation in the cytoplasm. Recently, a few reports have indicated that RAR and TR can shuttle from the cytoplasm to the nucleus in response to ligand (13–16). However, it is largely unknown which cellular factor(s) is responsible for the cytoplasmic retention of RAR and TR in the absence of their ligands.
In this study we identified a cytoplasmic retention factor and called it CART1; the protein has previously been called Rab11-FIP3, Arfophilin, and Eferin (17–19). Extensive binding assays indicated that CART1 interacted with helix H1 (D3 region) in the ligand binding domain (DEF or DE) of RAR or TR through its C-terminal conserved CoRNR box. As reported previously, NCoRs also interacted with a conserved helix H1 of RAR or TR through their two conserved CoRNRs (7, 31, 32). Here, we demonstrated that NCoR1 can compete with CART1 for RAR or TR binding, consistent with CART1 and NCoR being functionally related and sharing a common motif for RAR or TR binding.
What is the functional difference between CART1 and NCoR? NCoRs were originally identified as nuclear corepressors of RAR and TR in the absence of their ligands (7, 8). Our fluorescence microscopy revealed that CART1 is constitutively cytoplasmic and anchors RAR or TR in the cytoplasm in the absence of ligand but liberates RAR or TR to the nucleus in the presence of ligand. This ligand-induced shuttle from cytoplasm to nucleus gives rise to another aspect of the regulation of RAR or TR in addition to nuclear regulation mediated by NCoRs.
What is the consequence of the difference in the subcellular location of CART1 and NCoR? To address this, we used the fact that RAR functions as a transcriptional repressor by associating corepressor NCoRs in the nucleus in the absence of AtRA (37, 38). In CART1-deficient NIH3T3 cells, the transcriptional repression activity of RAR was evident but significantly abrogated by exogenous CART1 (Fig. 7A). Thus, CART1 could be a cytoplasmic derepressor of unliganded RAR by anchoring RAR repressor in the cytoplasm, thus preventing its nuclear translocation. This cytoplasmic checkpoint together with the nuclear checkpoint provided by NCoRs may be essential for the fine regulation of the transcriptional activity of RAR in the absence of ligand. CART1 also modulated the subcellular location and the transcriptional repression activity of TR (data not shown). In summary, CART1 appears similar to NCoRs with respect to selective NR (RAR and TR) binding using conserved binding motif and ligand utilization for binding but different from NCoRs in the subcellular location that leads to reverse transcriptional regulation in the absence of ligand.
What is the biological significance of the interaction between CART1 and RAR in the cytoplasm? To address this question, we used CART1-abundant TM4, one of the mouse testis Sertoli cell lines, where RAR was cytoplasmic in the absence of AtRA. CART1 knockdown in TM4 cells led to constitutive nuclear localization of RAR (Fig. 6D) and greatly increased repressing activity of RAR (Fig. 7D), likely due to more RAR interaction with NCoRs in the nucleus. Based on these observations, a possible role of CART1 in TM4 cells is postulated (Fig. 7E). Because CART1 is abundantly expressed in testis cell lines compared with the other mouse cell lines tested (Fig. 6C), we can speculate on the function of CART1 in testis development. Although ligand-induced RAR cytoplasm-to-nucleus shuttling has been reported in testis Sertoli cells (15, 16), little is known about the role of the putative cytoplasmic retention factor CART1 in testis. Recent data suggest that Rab11, another partner of CART1, is essential for fertility in the fruit fly (41). In contrast, vitamin A-deficient rat and RAR-null mouse models have established that RA and its receptor RAR are essential for early testis development (for review see Ref. 42).
Exclusive expression of CART1 in testis cell lines and its role in subcellular shuttling of RAR in TM4 Sertoli cells provide the possibility that CART1 is functionally linked to RAR in male reproduction. RAR regulation by CART1 in the cytoplasm may be an additional checkpoint for the proper functioning of RAR. Further molecular and genetic studies are needed to verify the physiological linkage between RAR and CART1 in the testis.
Supplementary Material
This work was supported by the Basic Science Research Program through NRF MEST Grants 2007-0053438 and 2008-0057731 (to S.-J. U.) and 2009-0064593 (to E.-J. K.), Republic of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- NR
- nuclear receptor
- RAR
- retinoic acid (RA) receptor
- AtRA
- all-trans retinoic acid
- TR
- cTR, chicken thyroid receptor
- T3
- thyroid hormone
- CART1
- cytoplasmic adaptor for RAR and TR
- NCoR
- nuclear receptor co-repressor
- SMRT
- silencing mediator for retinoid and thyroid hormone receptors
- Rab11-FIP
- Rab11-family interacting protein
- DBD
- DNA-binding domain
- AD
- activation domain
- HcRed
- red fluorescent protein
- ARF
- ADP-ribosylation factor
- IP
- immunoprecipitation
- TR
- thyroid receptor
- WB
- Western blotting.
REFERENCES
- 1.Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambon P. (1995) Recent Prog. Horm. Res. 50, 317–332 [DOI] [PubMed] [Google Scholar]
- 3.Yamashita S. (1998) Histol. Histopathol. 13, 255–270 [DOI] [PubMed] [Google Scholar]
- 4.DeFranco D. B. (1999) Cell. Biochem. Biophys. 30, 1–24 [DOI] [PubMed] [Google Scholar]
- 5.Hager G. L., Lim C. S., Elbi C., Baumann C. T. (2000) J. Steroid Biochem. Mol. Biol. 74, 249–254 [DOI] [PubMed] [Google Scholar]
- 6.Kumar S., Saradhi M., Chaturvedi N. K., Tyagi R. K. (2006) Mol. Cell. Endocrinol. 246, 147–156 [DOI] [PubMed] [Google Scholar]
- 7.Hörlein A. J., Näär A. M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Söderström M., Glass C. K., Rosenfeld M. G. (1995) Nature 377, 397–404 [DOI] [PubMed] [Google Scholar]
- 8.Chen J. D., Evans R. M. (1995) Nature 377, 454–457 [DOI] [PubMed] [Google Scholar]
- 9.Nagy L., Kao H. Y., Chakravarti D., Lin R. J., Hassig C. A., Ayer D. E., Schreiber S. L., Evans R. M. (1997) Cell 89, 373–380 [DOI] [PubMed] [Google Scholar]
- 10.Wen Y. D., Perissi V., Staszewski L. M., Yang W. M., Krones A., Glass C. K., Rosenfeld M. G., Seto E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7202–7207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass C. K., Rose D. W., Rosenfeld M. G. (1997) Curr. Opin. Cell Biol. 9, 222–232 [DOI] [PubMed] [Google Scholar]
- 12.McKenna N. J., Xu J., Nawaz Z., Tsai S. Y., Tsai M. J., O'Malley B. W. (1999) J. Steroid Biochem. Mol. Biol. 69, 3–12 [DOI] [PubMed] [Google Scholar]
- 13.Bunn C. F., Neidig J. A., Freidinger K. E., Stankiewicz T. A., Weaver B. S., McGrew J., Allison L. A. (2001) Mol. Endocrinol. 15, 512–533 [DOI] [PubMed] [Google Scholar]
- 14.Davis P. J., Davis F. B., Lin H. Y. (2008) Steroids 73, 1013–1017 [DOI] [PubMed] [Google Scholar]
- 15.Akmal K. M., Dufour J. M., Vo M., Higginson S., Kim K. H. (1998) Endocrinology 139, 1239–1248 [DOI] [PubMed] [Google Scholar]
- 16.Braun K. W., Tribley W. A., Griswold M. D., Kim K. H. (2000) J. Biol. Chem. 275, 4145–4151 [DOI] [PubMed] [Google Scholar]
- 17.Shin O. H., Ross A. H., Mihai I., Exton J. H. (1999) J. Biol. Chem. 274, 36609–36615 [DOI] [PubMed] [Google Scholar]
- 18.Prekeris R., Davies J. M., Scheller R. H. (2001) J. Biol. Chem. 276, 38966–38970 [DOI] [PubMed] [Google Scholar]
- 19.Hales C. M., Griner R., Hobdy-Henderson K. C., Dorn M. C., Hardy D., Kumar R., Navarre J., Chan E. K., Lapierre L. A., Goldenring J. R. (2001) J. Biol. Chem. 276, 39067–39075 [DOI] [PubMed] [Google Scholar]
- 20.Ullrich O., Reinsch S., Urbé S., Zerial M., Parton R. G. (1996) J. Cell. Biol. 135, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiba T., Koga H., Shin H. W., Kawasaki M., Kato R., Nakayama K., Wakatsuki S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15416–15421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eathiraj S., Mishra A., Prekeris R., Lambright D. G. (2006) J. Mol. Biol. 364, 121–135 [DOI] [PubMed] [Google Scholar]
- 23.Schonteich E., Pilli M., Simon G. C., Matern H. T., Junutula J. R., Sentz D., Holmes R. K., Prekeris R. (2007) Eur. J. Cell. Biol. 86, 417–431 [DOI] [PubMed] [Google Scholar]
- 24.Kim E. J., Park J. S., Um S. J. (2002) J. Biol. Chem. 277, 32020–32028 [DOI] [PubMed] [Google Scholar]
- 25.Cho Y. S., Kim E. J., Park U. H., Sin H. S., Um S. J. (2006) J. Biol. Chem. 281, 17588–17598 [DOI] [PubMed] [Google Scholar]
- 26.Park J. S., Kim E. J., Kwon H. J., Hwang E. S., Namkoong S. E., Um S. J. (2000) J. Biol. Chem. 275, 6764–6769 [DOI] [PubMed] [Google Scholar]
- 27.Kim E. J., Park J. S., Um S. J. (2003) Nucleic Acids Res. 31, 5356–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu X., Lazar M. A. (1999) Nature 402, 93–96 [DOI] [PubMed] [Google Scholar]
- 29.Perissi V., Staszewski L. M., McInerney E. M., Kurokawa R., Krones A., Rose D. W., Lambert M. H., Milburn M. V., Glass C. K., Rosenfeld M. G. (1999) Genes Dev. 13, 3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagy L., Kao H. Y., Love J. D., Li C., Banayo E., Gooch J. T., Krishna V., Chatterjee K., Evans R. M., Schwabe J. W. (1999) Genes Dev. 13, 3209–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurokawa R., Söderström M., Hörlein A., Halachmi S., Brown M., Rosenfeld M. G., Glass C. K. (1995) Nature 377, 451–454 [DOI] [PubMed] [Google Scholar]
- 32.Wurtz J. M., Bourguet W., Renaud J. P., Vivat V., Chambon P., Moras D., Gronemeyer H. (1996) Nat. Struct. Biol. 3, 87–94 [DOI] [PubMed] [Google Scholar]
- 33.Baumann C. T., Maruvada P., Hager G. L., Yen P. M. (2001) J. Biol. Chem. 276, 11237–11245 [DOI] [PubMed] [Google Scholar]
- 34.Mackem S., Baumann C. T., Hager G. L. (2001) J. Biol. Chem. 276, 45501–45504 [DOI] [PubMed] [Google Scholar]
- 35.Maruvada P., Baumann C. T., Hager G. L., Yen P. M. (2003) J. Biol. Chem. 278, 12425–12432 [DOI] [PubMed] [Google Scholar]
- 36.Damm K., Thompson C. C., Evans R. M. (1989) Nature 339, 593–597 [DOI] [PubMed] [Google Scholar]
- 37.Brent G. A., Dunn M. K., Harney J. W., Gulick T., Larsen P. R., Moore D. D. (1989) New Biol. 1, 329–336 [PubMed] [Google Scholar]
- 38.Baniahmad A., Köhne A. C., Renkawitz R. (1992) EMBO J. 11, 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei L. N. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 47–72 [DOI] [PubMed] [Google Scholar]
- 40.Moore J. M., Guy R. K. (2005) Mol. Cell. Proteomics 4, 475–482 [DOI] [PubMed] [Google Scholar]
- 41.Tiwari A. K., Alone D. P., Roy J. K. (2008) Cell. Biol. Int. 32, 1158–1168 [DOI] [PubMed] [Google Scholar]
- 42.Wolgemuth D. J., Chung S. S. (2007) Soc. Reprod. Fertil. Suppl. 63, 11–23 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.