Abstract
The DNA of most vertebrates is depleted in CpG dinucleotides, the target for DNA methylation. The remaining CpGs tend to cluster in regions referred to as CpG islands (CGI). CGI have been useful as marking functionally relevant epigenetic loci for genome studies. For example, CGI are enriched in the promoters of vertebrate genes and thought to play an important role in regulation. Currently, CGI are defined algorithmically as an observed-to-expected ratio (O/E) of CpG greater than 0.6, G+C content greater than 0.5, and usually but not necessarily greater than a certain length. Here we find that the current definition leaves out important CpG clusters associated with epigenetic marks, relevant to development and disease, and does not apply at all to nonvertabrate genomes. We propose an alternative Hidden Markov model-based approach that solves these problems. We fit our model to genomes from 30 species, and the results support a new epigenomic view toward the development of DNA methylation in species diversity and evolution. The O/E of CpG in islands and nonislands segregated closely phylogenetically and showed substantial loss in both groups in animals of greater complexity, while maintaining a nearly constant difference in CpG O/E between islands and nonisland compartments. Lists of CGI for some species are available at http://www.rafalab.org.
Introduction
CpG dinucleotides are the target for DNA methylation, a modification of cytosine heritable during cell division. The process of CpG loss is by CpG methylation, deamination, and failure of the repair system to identify a U-G pair. The remaining CpGs clustering in regions are referred to as CpG islands (CGI). Interest in CGI grew when it was demonstrated that they are enriched in the promoters of vertebrate genes (Bird 1986). CGI were also found at the 3′ ends and exonic regions of mammalian genes (Larsen et al. 1992). Interest has grown even more as many investigators have observed altered DNA methylation (DNAm) of CGI in development and cancer (Feinberg 2007). We recently demonstrated that CGI shores, defined as regions within 2000 bp but not inside CGI, are useful predictors for the location of tissue- or cancer-specific differentially methylated regions (DMRs) (Irizarry et al. 2009).
The formal definition of a CGI is a region of at least 200 bp, with GC content (proportion of Gs or Cs) greater than 50% and observed-to-expected CpG ratio (O/E) greater than 0.6 (Gardiner-Garden and Frommer 1987). A readily available list of CGI is available from the UCSC GenomeBrowser (Kent et al. 2002); the list which was derived using algorithms that search for regions satisfying the original definition of CGI. There are two reasons why this algorithmic definition, which has been a useful benchmark, needs to be modified.
First, data shown by Irizarry et al. (2009) motivate the need for a more flexible CGI definition since many DMRs not associated with CGI are nevertheless in the shores of CpG-enriched sequences. For example, one DMR reported by Irizarry et al. (2009) was within 1000 bp of a 1411-bp region that appears to be a CpG cluster. Furthermore, this region coincides with a gene promote, for CLSTN3. Despite coinciding with two functional elements associated with CGI, this region meets only two of the three criteria of the formal definition: O/E is only 0.5. Therefore, this region is not in the GenomeBrowser list of CGI. We found many such examples. Specifically, the CGI list, created with our method, covered 94% of the DMRs reported by Irizarry et al. (2009), a dramatic increase from the 65% covered by the GenomeBrowser CGI. Second, the algorithmic definition of CGI takes no account of species differences. Indeed, at least for Arabidopsis the prediction of methylation patterns based on CpG density does not apply (Cokus et al. 2008).
The current definition of CGI is somewhat arbitrary because the choice of the cutoffs has a great influence on what is considered an island. This choice was likely derived from exploratory data analysis (e.g., Fig. 1 in Gardiner-Garden and Frommer 1987), but neither a biological argument nor a formal statistical motivation was used. Alternative algorithmic definitions have been proposed. For example, Takai and Jones (2002) demonstrated that with slightly more stringent cutoffs, the enrichment for promoter regions of genes was not affected much, but most Alu-repetitive elements were excluded from the CGI list. However, by focusing on only promoters of known genes, we find this alternative definition results in even less sensitivity for other functional elements. Furthermore, the GenomeBrowser list appears to have been filtered to remove repeats, which is a viable solution that does not involve changing to a more restrictive definition.
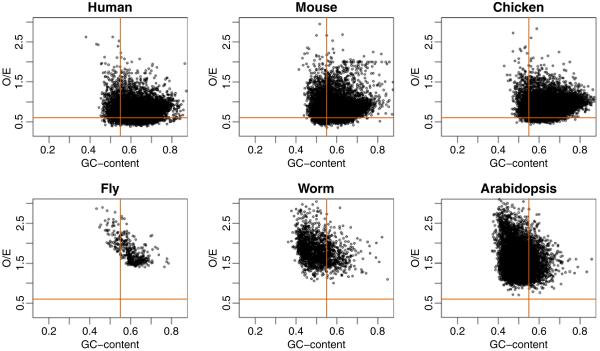
Fig. 1.
The O/E and GC content were computed for all islands found by our HMM procedure. O/E is plotted against GC content for six species. The vertical and horizontal lines represent the cutoffs used by the Gardiner-Garden and Frommer CGI definition
Glass et al. (2007) recently described a completely different algorithm based on the length of a segment needed to cover the nearest 27 CpGs. However, this 27-CpG requirement results in a list that leaves out many shorter CpG clusters that are associated with DMRs. For example, the cluster near CLSTN3, described above, is excluded. Furthermore, the Glass et al. approach, while valuable for analysis of human CGI, rejects the use of O/E of CpG as a criterion for island classification, although this enrichment over baseline may be an important reflection of the biological importance of these clusters or islands. The approach is also algorithmic and thus not necessarily applicable to interspecies comparison.
A general problem with existing algorithmic approaches is the need to specify thresholds (e.g., O/E > 0.6, number of CpG ≥ 27) that can be derived only from exploratory data analysis. Because the degree of CpG depletion varies across organisms, as we demonstrate below, the exploratory stage needs to be repeated to define CGI for other organisms. Because we assume that the underlying structure of the genome includes two unobserved states (CGI and baseline), Hidden Markov models (HMM) (Rabiner 1989) are a natural method to consider. A Markov process is a statistical model for the random movement of a system for which the probability of a given future state, at any given moment, depends only on its present state and not on any past states. In a HMM the system being modeled is assumed to be a Markov process but is not directly observed. The challenge is to determine the hidden parameters from observations that depend on these hidden parameters. In our scenario, the CGI and baseline regions are the hidden states and the CpG counts are the observations that depend on these states. Churchill (1989) introduced the use of HMM to sequence analysis. HHM have been previously used to model base-to-base transitions within a predetermined definition of CGI (Durbin et al. 1998). Such an approach, which is rather complicated, is not applicable to the genome-wide detection of CGI as it requires CGI to be predetermined for a training step. In contrast, we model CpG counts; this permits us to perform a totally data-driven procedure to identify CpG islands with no preconception regarding the properties of the genome. The statistical approach is described in the Methods section.
Methods
The mathematical details of our method are described elsewhere (Wu et al. 2009). Here we present a summary of the key points.
We followed the basic statistical concepts first used by Churchill (1989), described by Durbin et al. (1998), and used by bioinformatic tools such as MEME and BLAST. The foundation of these tools is the stochastic modeling of bases in the genome. We denoted B(t) the base at genomic location t, pb(t) the probability of B(t) = b for b = A, T, G, C, and pCG(t) the probability of being a CpG at location t. A useful model for detection of CGI needs two states to describe changes in pC(t), pG(t), and pCG(t). However, we have specified three parameters for each genomic location t, resulting in an over-determined system. To overcome this problem we modeled CpG counts in small intervals instead of the bases per se. This approach permitted the development of a tractable statistical model as described below.
We first divided the genome into nonoverlapping segments of length L (in bp). For the results shown here, we used L = 16. We denoted NC(s), NG(s), and NCG(s) as the number of C, G, and CpG in segment s, and Y(s) the hidden state for segment s with states Y(s) = 1 as CGI and Y(s) = 0 as baseline. We assumed, conditioned on pC(s) and pG(s) and Y(s) = i, a HMM model on NCG(s) with Poisson emission probabilities with conditional means:
Note that the parameters a1 and a0 can be interpreted as the O/E for the CGI (i = 1) and baseline (I = 0) regions, respectively. To simplify the model even further, we assumed that pC(s) = pG(s). The sense-antisense symmetry made this an acceptable assumption. We defined p(s) as the average {pC(s) + pG(s)}/2, which is equivalent to the expected GC content for segment s and rewrote the conditional means as
However, notice that this HMM model is over-parameterized as each segment s has a different conditional mean due to p(s). We solved this problem with a modular approach that estimated p(s) in a first step. We did this by assuming that conditioned on CGI state, p(s) was a smooth function of genomic location. We then fitted cubic splines to the observed GC content in segment s, {NC(s) + NG(s)}/L. With the estimate of p(s) in place, fitting the HMM to the CpG count data was achieved using standard algorithms. As a final step we obtained posterior probabilities of being in each state and created lists of CGI using different specificity cutoffs. Lists of CGI for some species are available at http://www.rafalab.org along with the posterior probabilities for the entire genome.
Results
We used an HMM approach applied to CpG counts that, unlike previous definitions, is data-driven rather than algorithmic and enabled us to preserve the underlying biological rationale of the Gardiner-Frommer approach of incorporating observed-to-expected CpG within the HMM, reasoning that a higher ratio is maintained by some evolutionary (functional) selection. Instead of fitting a model to the individual nucleotides, we fit a statistical model to the GC content and CpG counts in small bins (Wu et al. 2009). The model can be fitted without arbitrary cutoff choices, permits statistical testing for the presence of CGI, and provides a probability of being part of a CGI for any genomic region. Furthermore, the model parameters have relevant interpretations. For example, two parameters, which we denote as a0 and a1 (see Methods), represent the average O/E in the baseline and CGI regions, respectively.
With the model fit in place, each genomic location is assigned a posterior probability of being part of an island. An advantage of the probabilistic representation is that we can easily increase sensitivity while controlling specificity. For example, using a specificity level of 90% produced a list that covered 94% of the DMRs reported by Irizarry et al. (2009). The GenomeBrowser list covers 65%. We also tested the new lists on mouse DMR data (Yagi et al. 2008). A specificity level of 90% produced a list that covered 50% of the DMRs compared with the 20% with the GenomeBrowser. The differences in percentages between mouse and human data are likely due to differences in false-positive rates among the reported DMR lists. Note that different microarray protocols and statistical approaches were used by Irizarry et al. (2009) and Yagi et al. (2008).
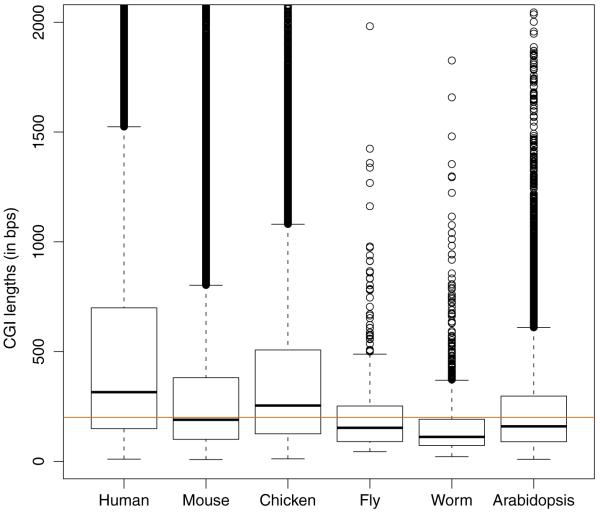
To understand the characteristics of the CGI in our list but not included in the GenomeBrowser list, we plotted GC content versus O/E for the model-based CGI (Fig. 1). The horizontal and vertical lines are from the Gardiner-Garden and Frommer CGI definition (GC content > 50%, O/E > 0.6). Based on the current definition, only the points above the horizontal line and to the right of the vertical line are CGI. Many of the model-based CGI do not satisfy the original definition. Furthermore, a box-plot of the lengths of model-based CGI shows that many model-based islands are smaller than the formal definition’s requirement of 200 bp (Fig. 2). Remarkably, though, the average length of CGI across species is quite similar, although the variance across species varies (human high, worm low).
Fig. 2.
Lengths (in bases) for the islands of each species displayed in Fig. 1. The horizontal line represents the cutoff used by the Gardiner-Garden and Frommer CGI definition
Where our HMM approach showed the greatest utility was in the comparative analysis of CpG distribution across species. We fitted our model to 30 species: E. coli, yeast, Arabidopsis, rice, C. elegans, C. japonica, C. brenneri, C. remanei, C. briggsiae, D. melanogaster, D. simulans, D. yakuba, zebrafish, fugu, stickleback, medakafish, tetraodon, chicken, zebra finch, cow, dog, mouse, rat, horse, opossum, human, chimpanzee, orangutan, honeybee, and mosquito. We tested for the presence of CGI by computing a likelihood ratio comparing a model with two states to a model with one state. Of the 30 species we tested, only the unicellular organisms, i.e., yeast and E. coli, did not show significant evidence in favor of the presence of CGI. Figure 3 shows the O/E of CGI and baseline (non-CGI) for each of the 30 species. Note the strong clustering within phylogenetic groups. Such an analysis has not been possible previously since the conventional algorithmic CGI definition provides lists for only mammals and birds (see GenomeBrowser).
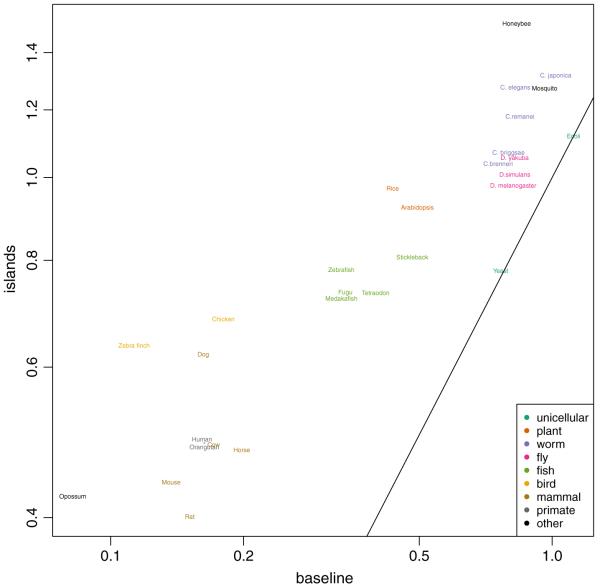
Fig. 3.
a1 (O/E for islands) plotted against a0 (O/E for baseline) for 30 species. Colors are used to define phylogenetic groups. Points on the identity line represent species with no islands. The axes are in the log scale. Chimpanzee is denoted with an asterisk to avoid clumping of names
The baseline O/E is depleted across all of the species, to varying degrees, but much more so in warm-blooded animals, with the curious exception of the marsupial Monodelphis, which is the most depleted of all. The vertebrates were CpG depleted in the baseline state as shown by estimates of a0 below 0.2, but for Monodelphis a0 was 0.08. In contrast, for fish the estimate of a0 was approximately 0.4, and for worms and insects a0 was approximately 0.9.
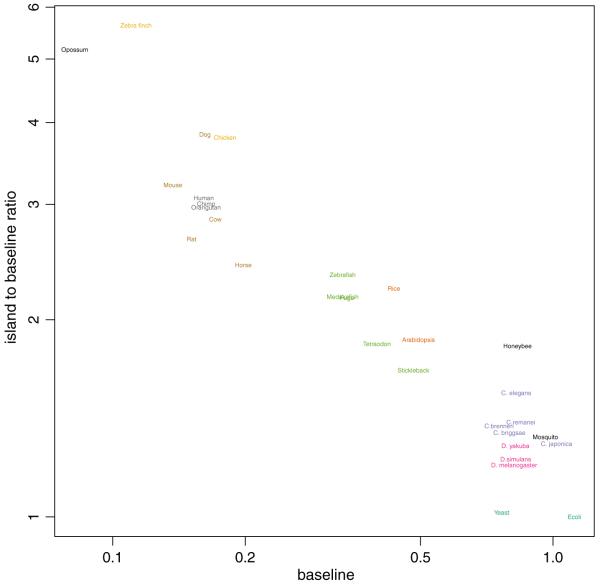
For most species the estimates of a1 were about 0.3 higher than a0. The estimates of a1 ranged from 38% larger (worm) to almost twice as large (bee). A more subtle point from inspection of the data is that the rate of decline of a1 is less than that of a0 as it progresses to a lower a0. This can be seen more clearly when the a1/a0 ratio is plotted against a0 on a log-log plot (Fig. 4), showing a linear relationship between the ratio and a0 in the log scale. These data suggest that both CGI and non-CGI lose CpG, presumably because of the higher mutation rate at methylated cytosine, but are under differing evolutionary pressure for their retention.
Fig. 4.
As Fig. 3 but the a1/a0 ratio is plotted on the y axis instead of a1
Discussion
In summary, we have made two significant advances in understanding the relationship between CGI and comparative genomics. The first is that using a new model-based definition, one can observe CGI across all multicellular organisms that have been sequenced, and they clearly do not fall within conventional algorithmic definitions such as the Gardiner-Frommer classification. Even the human model-based CGI list differs from the classical definition and includes greater coverage of the CGI whose shores show differential methylation across tissues and in cancer. Note, that evidence of methylation has been reported for species for which we found evidence of CGI. The flies had the weakest evidence for the presence of CGI, although it was still statistically significant. This would argue that there is some small component of methylation in flies. Interestingly, small amounts of methylation are detected for this organism (Lyko et al. 2000). The bee, in contrast, shows a marked enrichment of the CGI compartment, even though these very CGI would not be predicted by a conventional algorithmic definition. This result is consistent with recent findings showing the importance of DNA methylation in social caste development (Kucharski et al. 2008) and an association with development among genes near CpG-rich regions (Elango et al. 2009). Note that CpG depletion in the species studied here may not be entirely due to DNA methylation as CpG depletion also occurs in some bacteria, mitochondria, and viruses that do not have CpG methylation.
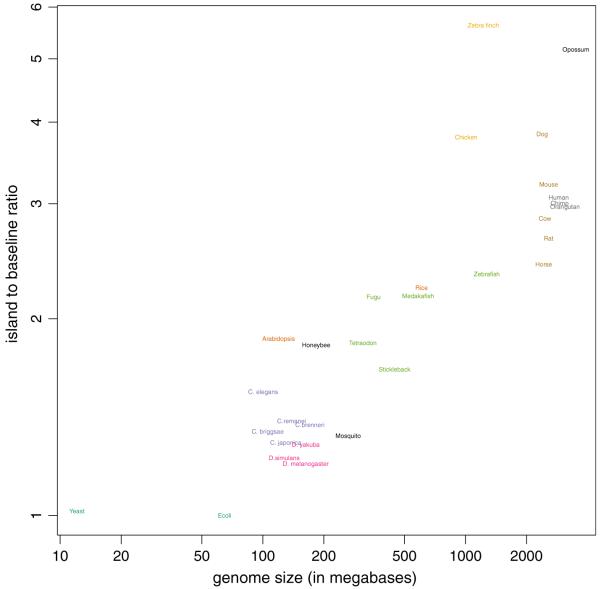
The second significant advance of this work is that it allows for the first time comparative functional analysis of evolutionary epigenetics across all of the multicellular organisms that have been sequenced. The new approach facilitated the generation of CGI for other species and we fitted the model to the genome of 30 species. This led to some interesting findings. Strong evidence for the presence of CGI was found for all multicellular organisms examined. This epigenetic classification shows striking segregation of phylogenetic categories: CpGs are progressively depleted from insects to worms, to fish, to plants, to birds, to mammals, and to the one marsupial represented. The rate of depletion of CpGs within CGI along this progression is less than outside CGI, but close to constant. This is most consistent with the idea of a greater degree of CpG loss with organismal complexity. This can easily be seen if we quantify complexity with genome size (Fig. 5). In addition, there are notable exceptions such as the differences across insects and mammals, with the honeybee and dog showing a much higher a1/a0 than related organisms, and more subtly for humans compared to other primates (see Fig. 4). This suggests a greater selective pressure for maintaining CpG within CGI in those species.
Fig. 5.
The a1/a0 ratio is plotted against genome length
Acknowledgments
NIH grants P50HG003233 and R01GM083084 supported this work. We also thank Harris Jaffee and Brian Caffo for their input and the reviewers for useful comments that reshaped the manusctipt.
Contributor Information
Rafael A. Irizarry, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, 615 North Wolfe Street, E3620, Baltimore, MD 21205, USA
Hao Wu, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, 615 North Wolfe Street, E3620, Baltimore, MD 21205, USA.
Andrew P. Feinberg, Center for Epigenetics, Johns Hopkins University School of Medicine, 855 N. Wolfe St., Rangos 570, Baltimore, MD 21205, USA
References
- Bird A. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Churchill GA. Stochastic models for heterogeneous DNA sequences. Bull Math Biol. 1989;51:79–94. doi: 10.1007/BF02458837. [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R, Eddy SR, Krogh A, Mitchison G. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press; Cambridge, UK: 1998. [Google Scholar]
- Elango N, Hunt BG, Goodisman MA, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci U S A. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Glass J, Thompson RF, Khulan B, Figueroa ME, Olivier EN, et al. CG dinucleotide clustering is a species-specific property of the genome. Nucleic Acids Res. 2007;35:6798–6807. doi: 10.1093/nar/gkm489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, et al. Genome-wide methylation analysis of human colon cancer reveals similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):246–250. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Lyko F, Ramsahoye B, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408:538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- Rabiner L. A tutorial on hidden Markov models and selected applications in speech recognition. Proc IEEE. 1989;77:257–286. [Google Scholar]
- Takai D, Jones P. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Caffo B, Jaffee HA, Feinberg AP, Irizarry RA. Redefining CpG Islands using a Hideen Markov model. Johns Hopkins University, Dept. of Biostatistics Working Papers; 2009. Working Paper 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, et al. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 2008;18:1969–1978. doi: 10.1101/gr.074070.107. [DOI] [PMC free article] [PubMed] [Google Scholar]