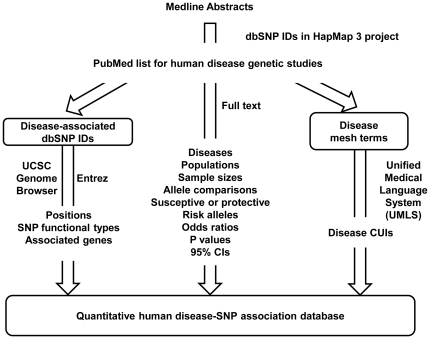
Figure 1. A curated quantitative disease-SNP association database.
Starting from a list of all SNPs measured in the HapMap 3 project, we searched for their presence in all Medline abstracts, eliminating non-human studies. Significant SNP-disease associations were manually curated from the full text, and reviewed four rounds. SNP IDs were annotated using the UCSC genome browser for positions and function types and annotated using Entrez for associated genes. Disease mesh terms were compared with the Unified Medical Language System (UMLS) to select concept unique identifiers (CUIs).