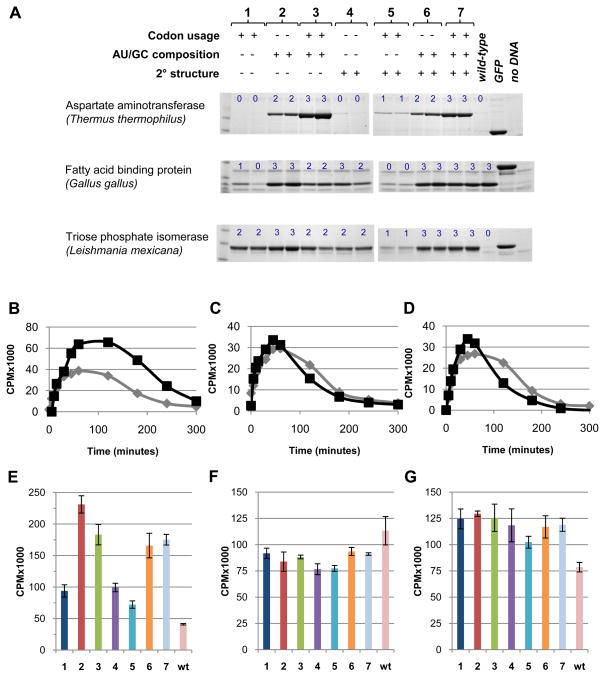
Figure 2. Experimental expression levels of synthetic genes determined using E. coli coupled in vitro transcription and translation reactions.
(A) Synthetic genes were designed by optimizing CAI, mRNA secondary structure, and 5′ ORF regional nucleotide composition singly or in combination giving a total of seven conditions. For each condition, the expression pattern of two alleles differing by at least 10 mutations are shown. Three proteins differing in size, structure, origin and expression of wild-type ORF sequences were used: asparate aminotransferase (ttAST), fatty acid binding protein (ggFABP), and triose phosphate isomerase (lmTIM). Proteins were purified from coupled in vitro transcription and translation (TnT) reactions using immobilized metal affinity chromatography and run on 4–12% SDS-PAGE gradient gels. Green florescent protein template was included as a positive control for protein expression levels and an extract without added DNA as a negative control. Observed expression levels were classified into one of four categories (blue numbers: 0, no band; 1, weak band; 2, medium band; 3, strong band). Full gel images are shown in Figure S1. The identity of the observed protein band was verified by mass spectrometry for each of the three proteins in the first allele of the optimization condition 7 (Figure S2). (B–D) Time course of radiolabeled RNA in TnT reactions containing a high- (black) and low- (grey) expression level allele (background of a reaction without added DNA was substracted): ttAST (B), ggFABP (C), lmTIM (D). (E–G) Total radiolabeled RNA at one hour using one allele for each condition presented in panel A and the wild-type sequences: ttAST (E), ggFABP (F), lmTIM (G).