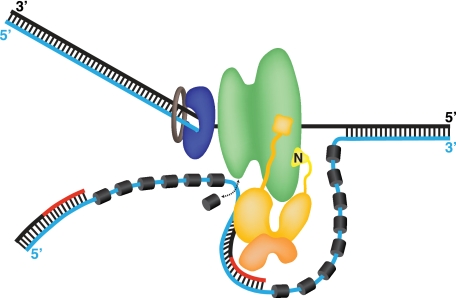
FIGURE 7.
Speculative model of the SV40 primosome at a replication fork. A single Tag hexamer (green) tracking 3′–5′ on the leading strand template (black) is followed by DNA polymerase δ (blue)-proliferating cell nuclear antigen (brown) holoenzyme. The lagging strand template is displaced as Tag unwinds the duplex, but its point of exit from the helicase and its path after exit are controversial (17–20, 42, 43). Hence, the path depicted here is the simplest possibility. RPA (dark gray cylinders) bound to the lagging strand template (cyan line) is remodeled by specific contacts with the OBDs of the Tag hexamer into a weaker ssDNA-binding mode (22, 23, 35) that is easily displaced (arrow), exposing the template. The p180/p68 (gold) subunits of pol-prim contact the helicase domains of Tag through p68N (36) (see Figs. 1–3) and the N terminus of p180, which we propose are flexibly tethered to the pol-prim complex (27, 29, 30). The primase p58/p48 (orange) interaction surfaces with Tag are not known (30). Pol-prim correctly positioned on Tag is postulated to access the template exposed upon RPA remodeling, synthesize an RNA primer (red line), and extend it (black line), yielding an RNA-DNA primer. The subsequent proliferating cell nuclear antigen clamp loading and switch to DNA polymerase δ holoenzyme are not depicted (34, 71).