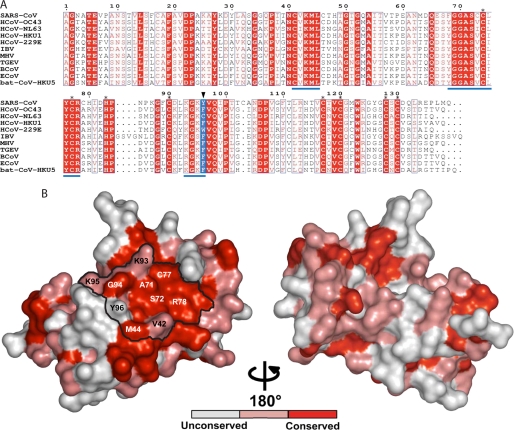
FIGURE 2.
nsp10 protein conservation among coronavirus groups 1, 2, and 3. A, sequence alignment of nsp10 proteins derived from genome sequences of the following: SARS-CoV, SARS coronavirus (group 2b, NC_004718), HCoV-OC43, human coronavirus OC43 (group 2a, NC_005147), HCoV-NL63, human coronavirus NL63 (group 1, NC_005831), HCoV-HKU1, human coronavirus HKU1 (group 2a, NC_006577), HCoV-229E, human coronavirus 229E (group 1, NC_002645), IBV, infectious bronchitis virus (group 3a, NC_001451), MHV, mouse hepatitis virus (group 2a, NC_006852), TGEV, transmissible gastroenteritis virus (group 1, NC_002306), BCoV, bovine coronavirus (group 2a, NC_003045), ECoV, equine coronavirus (group 2a, NC_010327), and bat coronavirus, bat-CoV-HKU5–1 (group 2c, NC_009020). The black triangle points to residue 96, colored in blue. The sequences were aligned using the ESPript program (46). Stars indicate residues involved in coordinating zinc atoms. Underlined sequences in blue correspond to clusters of mutations obtained by RY2H. B, nsp10 from SARS-CoV (Protein Data Bank code 2FYG) is depicted in a surface representation. From the alignment above, absolutely conserved residues are shown in red, whereas conserved residues and non-conserved residues are shown in pink and white, respectively. The different amino acids identified as being part of nsp10-nsp16 interaction are named, and the potential surface of interaction is delineated in black.