FIGURE 6.
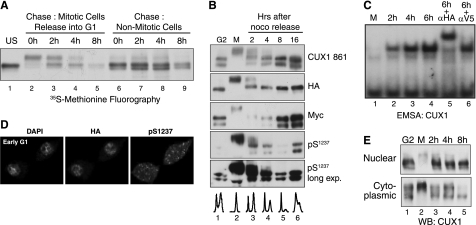
Phosphorylation-dephosphorylation restores low CUX1 DNA binding in G1. A, pulse-chase analysis in T98G cells expressing a recombinant p110 CUX1 protein (Myc/747-1505/HA). Cells were incubated for 10 h in the presence of nocodazole and [35S]methionine; floating (Mitotic) and attached (Nonmitotic) cells were separated and re-fed with fresh medium. At the indicated time points, whole cell extracts were prepared and submitted to immunoprecipitation with anti-CUX1 antibodies; proteins were separated by SDS-PAGE and revealed by fluorography. Only attached cells were collected for lanes 6–9 to look at CUX1 stability in nonmitotic cells. B, T98G cells expressing p110 CUX1(Myc/747-1505/HA) were treated with nocodazole for 12 h; attached (G2) and floating (M) cells were separated, and the floating cells were replated in fresh medium; at the indicated time, whole cell extracts were prepared and analyzed by immunoblotting with the indicated antibodies using amounts of protein lysates as specified in B. C, U2OS cells expressing p110 CUX1(Myc/747-1505/HA) were treated with nocodazole for 12 h, and the floating cells were replated in fresh medium. At the indicated time points, extracts were prepared and analyzed in EMSA using a CUX1 probe, as described in Fig. 2B. D, cells were prepared as in C but were replated on a cover slide, incubated for 4 h, and then stained with DAPI and analyzed by indirect immunofluorescence using HA and Ser(P)1237-specific antibodies. E, T98G expressing p110 CUX1 was treated as described in C, and subcellular fractionations were performed to obtain crude cytoplasmic and nuclear extracts, which were analyzed by immunoblotting using anti-CUX1 antibodies.