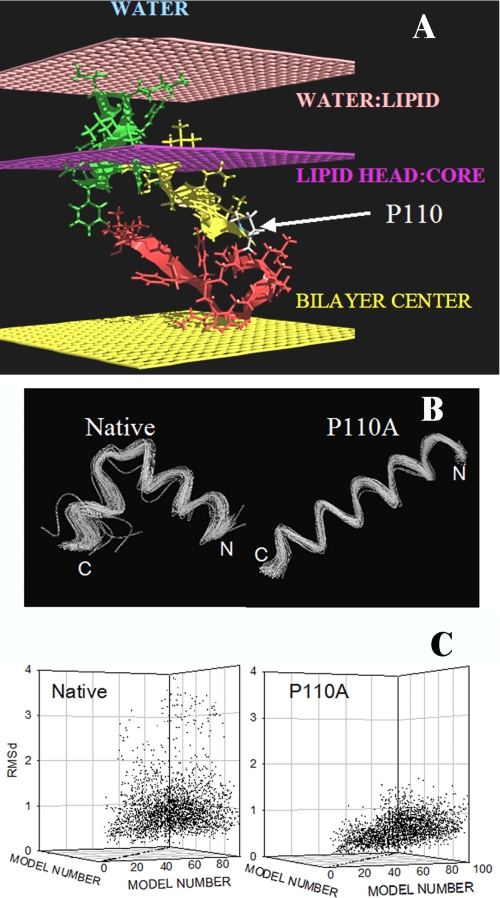
FIGURE 7.
Modeling of the transmembrane segment of caveolin-1 using PepLook. A, best position of the caveolin 94–122 fragment in a membrane. All PepLook models were systematically pulled through a membrane slab bilayer (50), across the water/lipid interfaces (light pink layers), the lipid polar heads/acyl chain interfaces (dark pink layers), and the membrane center (yellow layer). The model structure and position corresponding to the lower restraint energy is here presented. This model has the U bent conformation; the N- side is yellow, and the C-side is red. The center is in the hydrophobic layer of membrane, and the 94VTKYWFYRL102 fragment (green) is in the membrane interface layer in connection with lipid polar heads. Note that only one monolayer of the membrane bilayer is shown in the figure. The position of the proline residue is indicated. B, spline image of the 99 three-dimensional models calculated using PepLook (49). All models are fitted by adjusting their backbone atoms (N–C–C=O) to those of the lowest energy structure (the Prime), the three-dimensional model of lower energy using Qmol. The N- and C-ends are indicated. Shown are conformations of the 99 PepLook three-dimensional models of the caveolin fragment 103–122. Left, native fragment; right, P110A mutant. C, r.m.s. deviation matrices of all pairs of three-dimensional models.