Abstract
Background
Similar to a subset of human patients who progress from monoclonal B lymphocytosis (MBL) to chronic lymphocytic leukemia (CLL), New Zealand Black (NZB) mice have an age-associated progression to CLL. The murine disease is linked to a genetic abnormality in microRNA mir-15a/16-1 locus, resulting in decreased mature miR-15a/16.
Methods
Spleens of aging NZB were analyzed for the presence of B-1 cells via flow cytometry and for the presence of a side population (SP) via the ability of cells to exclude Hoechst 33342 dye. The SP was assayed for the presence of hyperdiploid B-1 clones and for the ability to differentiate into B-1 cells in vitro and transfer disease in vivo. In addition, enhanced apoptosis of chemoresistant NZB B-1 cells was examined by restoring miR-16 levels in nutlin-treated cells.
Results
Aging NZB mice develop a B-1 expansion and clonal development that evolves from MBL into CLL. An expansion in SP is also seen. Although the SP did contain increased cells with stem cell markers, they lacked malignant B-1 cells and did not transfer disease in vivo. Similar to B-1 cells, splenic NZB SP also has decreased miR-15a/16 when compared with C57Bl/6. Exogenous addition of miR-15a/16 to NZB B-1 cells resulted in increased sensitivity to nutlin.
Conclusion
NZB serve as an excellent model for studying the development and progression of age-associated CLL. NZB SP cells do not seem to contain cancer stem cells, but rather the B-1 stem cell. NZB B-1 chemoresistance may be related to reduced miR-15a/16 expression.
Keywords: chronic lymphocytic leukemia, monoclonal B cell lymphocytosis, miR-16, murine model of CLL, New Zealand Black, side population
Chronic lymphocytic leukemia (CLL) is an age-associated malignancy of the CD5+ B cell (1–3). Although CLL is one of the most common leukemias in the Western Hemisphere (4), the etiology of this disease is not fully understood. As with most cancers, no single genetic aberration or transformation event has been identified with the initial onset of CLL. Despite familial associations (2) and the presence of stereotyped sequences on B-cell receptor immunoglobulin (5), the genetic basis for development of CLL remains unresolved.
Genomic “fragile sites” harboring microRNAs have been implicated in the development of cancers (6). Cytogenetic abnormalities associated with CLL include chromosomal duplications and deletions, particularly the 13q14 chromosomal region (7) containing the DLEU2 and mir-15a/16-1 genes (8,9), seen in over 50% of patients. Alterations in this genomic region containing microRNAs, mir-15a and mir-16-1 are present in a sub-population of B-CLL patients (10,11). Family members of patients with CLL have an increased chance of developing the disease (2). Some family members of CLL patients have also been found to harbor B cells with immunophenotypes very similar to CLL B cells, though not displaying symptoms of disease (12). Evidence suggests that CLL is preceded by monoclonal B-cell lymphocytosis (MBL), a lymphoproliferative disorder characterized by CD19+ B cells expressing CD5/CD20/CD79b in the absence of marked symptoms of hematologic disease (13–15).
Typical MBL phenotype is detected in a subset of healthy first-degree relatives of CLL patients, indicative of an inherited predisposition (12). Although most CLL cases demonstrate a single dominant clone, it is unclear whether MBL cases are pauciclonal or monoclonal, as its misleading name suggests. In a recent study by Lanasa et al., four of six MBL cases consisted of two or more unrelated clones, as well as 13q14 deletions, suggesting an early involvement of miR-15a/16 in the progression to CLL (16).
The New Zealand Black (NZB) mouse model is a de novo model of CLL (17), in contrast to all other models, which are induced by the expression of exogenous genes (18). Similar to CLL, the disease in NZB mice is an age-associated malignant expansion of poly-reactive CD5+ B-1 clones (5,18). The majority of B-1 clones are IgM+, B220 (CD45R)dim and CD5dim, increase with age, and often possess chromosomal abnormalities (19). NZB also seem to demonstrate an MBL-like stage at an early age, characterized by multiple clones, as seen in MBL cases reported by Lanasa et al. (16). High levels of IL-10 are also correlated with the development of these malignant B-1 cells (20). This MBL-like state in NZB precedes CLL, and although it exhibits similar manifestations to human MBL, NZB disease will always progress to CLL, in contrast to humans who can have an indefinite state of indolent MBL disease (16). The NZB has also been studied as a model for autoimmunity (21). Similar to the autoreactivity associated with CLL autoantibodies (22), the NZB displays a mild autoimmunity associated with B cell hyperactivity, resulting in autoimmune hemolytic anemia (AIHA) and antinuclear antibodies (18).
We have previously found the development of the NZB disease to be associated with a germline genetic alteration in the mir-15a/16-1 locus, which is correlated with a decrease in mature miR-15a and miR-16 expression in lymphoid tissues (23). The NZB exhibits a T→A point mutation six bases downstream from pre-mir-16-1 on mouse chromosome 14 (23), similar to the C→T point mutation seen in human CLL on human chromosome 13 (24), which may affect structural stability of the stem loop and proper processing to mature form. This latter mutation is a rare event and has only been reported in two human CLL patients. In the majority of human CLL patients, the deletion of the 13q14 region results in a loss of the mir-15a/16-1 locus. Both genetic alterations result in a decrease in mature miR-15a and miR-16. All NZB mice exhibit the germline point mutation in mir-15a/16-1 rather than a chromosomal deletion, and this mutation is associated with decreased levels of miR-15a/16 (23). Decreased expression of miR-16 has been shown to play a role in deregulated cell cycle transit, resulting in G1 accumulation upon restoration (25–27) aided by the direct targeting of, and reduction in, cyclin D1 protein levels (27). Restoration of miR-16 levels to an NZB-derived malignant B-1 cell line was also shown to enhance drug sensitivity, synergizing with nutlin, an MDM2 antagonist and p53 activator (28), and with genistein, a tyrosine kinase inhibitor (29), augmenting apoptosis (27).
Side populations (SP) are small groups of primitive, undifferentiated cells (30) that possess the ability to exclude Hoechst 33342 dye via a multidrug resistance protein (MDR) (31,32). Studies have demonstrated tumors to be composed of a heterogeneous population of cells, in which only a small subset exhibits the capability of reconstituting a tumor when injected into mice and serially transplanted (30,33). We found the percentage of cells in the SP from old NZB to be dramatically elevated relative to young NZB and to young or old non-NZB strains, and to also display surface markers characteristic of stem cells (c-kit, sca-1) (34). In the present report, we analyzed the characteristics of sorted SP cells to determine the presence of a unique microRNA signature and analyzed this population for the presence of B stem cells and CLL progenitor cells. Our data further support the unique advantage the NZB strain offers for following the natural history of the age-associated development of CLL from B-cell hyperactivity and MBL-like disease.
METHODS
Sources of Cells
Mice: NZB/BlNJ, C57Bl/6J, and DBA/2J were obtained from Jackson Laboratories (Bar Harbor, ME). The NZB derived B-1 cell line, LNC, was derived from a year-old NZB as previously described (35).
Analysis of DNA Content
Single-cell suspensions isolated from NZB spleens were stained with hypotonic propidium iodide (PI) (0.05 mg/ml PI, 0.1% Triton X-100, 0.1% sodium citrate) and acquired on Becton Dickinson FACSCalibur using CELLQUEST software (Becton Dickinson) and analyzed using ModFit LT V3.1 software (Verity Software House, Topsham, ME).
Detection of Malignant B-1 Cells
Splenic single cell suspensions were obtained from 3-, 9-, and 15-month-old NZB and C57Bl/6 mice. For detection of malignant B-1 cells, cells were stained with FITC anti-IgM antibody (Ab) (Caltag Invitrogen), Tri-Color anti-B220 (CD45R) Ab (Caltag Invitrogen) and PE-anti-CD5 Ab (Pharmingen San Diego, CA). Cells were then fixed in 2% paraformaldehyde. All samples were run on a FACSCalibur (BD Biosciences, San Jose, CA), collecting data on 2 × 104 events and analyzed using CellQuest Software (BD Biosciences, San Jose, CA), gating on the lymphoid population based on forward and side scatter.
Cell Sorting of Spleen Cells
Spleens were removed from 9-to 14-month-old C57Bl/6 (control non-NZB strain) and NZB mice and made into single-cell suspensions that were four-color stained with antibodies directed against IgM fluorescein isothiocyanate (FITC), B220 PerCP-Cy5.5, CD4 phycoerythrin (PE), and CD25 allophycocyanin (APC) (Invitrogen, Carlsbad, CA) and sorted into four distinct populations on a FACSAria (Becton Dickinson, San Jose, CA). The B cells were gated on CD4− and sorted into two populations (B-2 cells: CD4−/IgM+/B220bright and B-1 cells: CD4−/IgM+/B220dim).
MicroRNA Extraction and Quantification
Total RNA, including miRNA, was extracted from B-1 and B-2 subpopulations sorted from spleens of NZB and normal strain mice according to the Trizol (Invitrogen) manufacturer's protocol. Quantitative real time PCR (qPCR) was used to quantify mature miR-16 expression using the TaqMan microRNA Reverse Transcription and TaqMan miRNA hsa-miR-16 Assay Kit (Applied Biosystems, Foster City, CA). The qPCR reaction was run on the Applied Biosystems 7500 Real-Time PCR Systems for 40 cycles at 60°C. The relative quantification (RQ) values of NZB miR-16 levels compared with normal mouse strain miR-16 levels were determined using the standard 2−ΔΔCT method according to the manufacturer's protocol, comparing the difference in the amount of miR-16 and U6 in the calibrated control sample from the difference in the amount of miR-16 and U6 in the NZB sample. The total amount of input RNA was normalized to the Taqman U6 snRNA housekeeping gene (Applied Biosystems).
Detection of Apoptosis in Nutlin-Treated Cells
A lentiviral sensor assay was performed as previously described (27), employing a bidirectional lentiviral vector containing cyclin D1 3′UTR miR-16 binding sites downstream of the green fluorescence protein (GFP) cassette, to detect the activity of exogenously transfected miR-16. The lentiviral vector also expresses monomeric cherry (mCherry) as an internal control. The transduced NZB cell line LNC was then transfected with 100 nM miR-16 or negative control (Dharmacon) and treated with 5 μM nutlin-3 (Sigma) as previously described (27). Cells were plated at 0.5 × 106 cells/ml and incubated at 37°C for 48 hours. For detection of apoptosis, cells were incubated in the dark for 15 min at 4°C in 5 μl annexin V PE (Invitrogen), according to manufacturer's protocol. Samples were washed with 1 × binding buffer and acquired within 1 hour. Five thousand events per sample were acquired on the Amnis ImageStream IS100 and analyzed for GFP, mCherry, and annexin V expression using the Amnis IDEAS 3.0 software (Amnis Corporation, Seattle, WA).
Isolation of “Side Population” (SP) by Flow Cytometry
Cells from spleens and bone marrow of NZB, DBA/2, and C57Bl/6 mice were isolated and subjected to RBC lysis, using RBC Lysis Solution (Gentra Puregene, Qiagen, Valencia, CA) according to the manufacturer's protocol. One to two million cells were incubated at 37°C in the presence of Hoechst 33342 (5 μg/ml) with or without coincubation with Verapamil (50 μM), an ABC transporter inhibitor which blocks Hoechst exclusion from the cells and eliminates SP visibility, for 90 minutes as previously described (32). For those SP analyzed for surface markers, after Hoechst incubation, cells were then stained with sca-1 PE, B220 FITC, CD19 APC, and c-kit APC. For the detection of dead cells, 2 μg/ml propidium iodide in PBS was added. Samples were acquired on the LSRII (BD Biosciences) exciting Hoechst with the 355nm UV laser, using the 670/LP for detection of Hoechst red and 450/50 bandpass filter to detect Hoechst blue. The 630/22 bandpass filter was used for the detection of PI excited by the blue laser. Acquisition and analysis were performed using FACSDiva Software 6.0 (BD Biosciences). Detection of a SP was verified by its absence in the presence of Verapamil (32), gating on the live PI negative population.
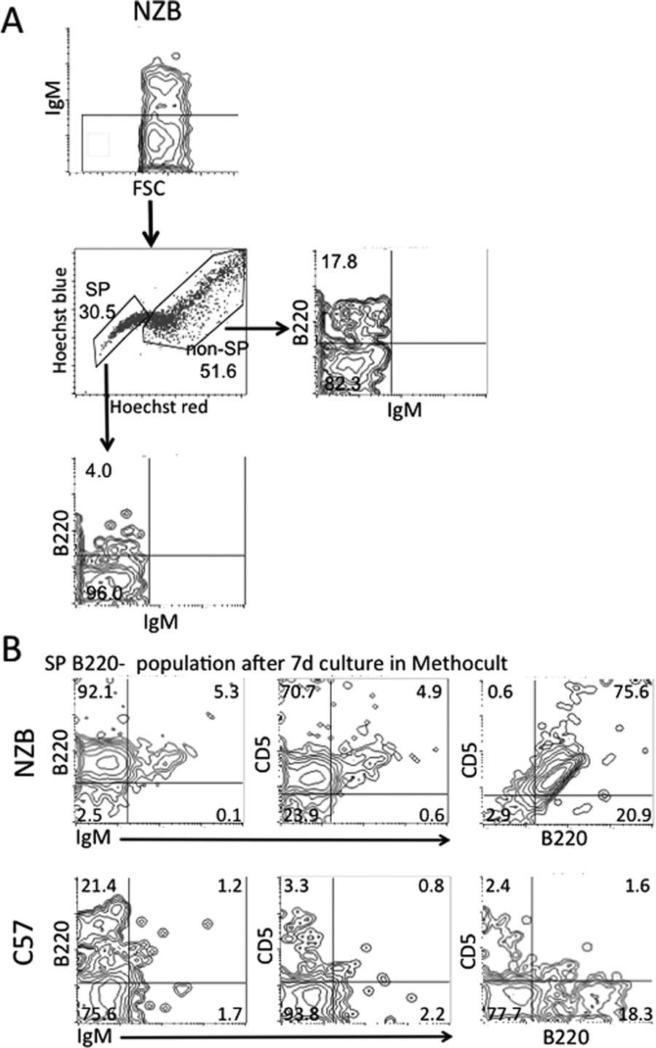
Colony-Forming Unit (CFU)-Pre-B Cell Assay on Sorted SP and Non-SP
Single cell suspensions were obtained from an enlarged spleen of a 17-month-old NZB and from 8- to 10-month-old C57Bl/6 mice (pooled three spleens). Cells were stained for SP using the aforementioned procedure. Following staining for the SP, cells were stained with B220 (CD45R) FITC, IgM PE, and 7-aminoactinomycin D (AAD). Cells were then gated on 7AAD−/IgM− and sorted on a FACSVantage Cell Sorter into the following populations: SP B220− , SP B220+, nonSP B220−, and nonSP B220+. Each of the four sorted populations were then cultured in duplicate in Methocult M3630 methycellulose medium for mouse pre-B cell colonies (Stem-Cell Technologies, Vancouver, BC, Canada), according to the manufacturer's protocol and incubated at 37°C for 7 days. On the seventh day, colonies were counted using a light microscope and classified on the basis of size (small having 10–20 cells per colony, medium 20–30, large 30–40, and very large >50 cells). Cells were then harvested from the methycellulose medium according to the manufacturer's protocol, stained with IgM FITC or APC (Caltag), CD5 PE (BD Pharmingen), and B220 TC (Invitrogen), acquired on a FACSCalibur and analyzed using CellQuest Software.
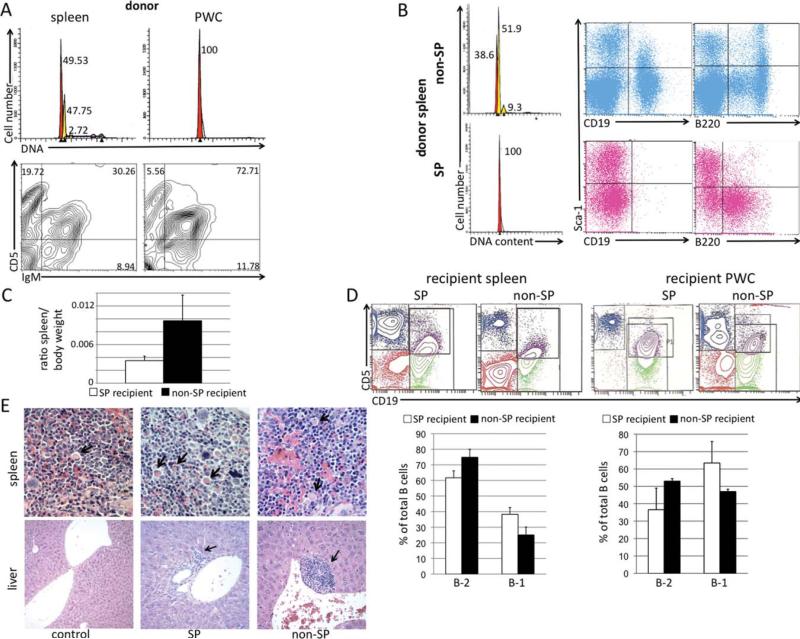
In Vivo SP/Non-SP Transfer
Splenic cells from a 13-month-old female NZB with disease were incubated in the presence of Hoechst 33342 as aforementioned and sorted into SP and non-SP cell groups. Two million SP or non-SP cells in a total volume of 200 μl were injected intraperitoneally into 4-week-old male NZB recipients (three in each group) and a control group of NZB were injected with 200 μl of syngenic spleen cells (three in each group). Mice were analyzed 5.5 months postinjection for hyperdiploidy by staining with hypotonic PI, and for the presence of malignant B-1 cells, staining with IgM or CD19 FITC, CD5 PE, B220 TC. Mice were also subjected to ultrasound imaging and histological analysis was performed on tissues.
Histopathology
Tissue sections for microscopic evaluation were made from paraffin-embedded spleen, kidney, and liver of mice injected with SP and non-SP and stained with hematoxylin and eosin. Tissues were analyzed for architecture, lymphocyte infiltration, and number and size of germinal centers using an Olympus BX40 microscope and 10x, 20x and 40x objective lenses. Digital photographs were taken.
Ultrasound Analysis of Spleen Size
Ultrasound imaging was performed at the NIH Mouse Imaging Facility (MIF) on NZB 3.5mo (n = 4), 7mo (n = 9), 10.5mo (n = 8), and 16mo (n = 7). Mice were anesthetized by continuous inhalation of 2-3% isoflurane with 1% oxygen and imaged with the VisualSonics Vevo™ 770 In Vivo High-Resolution Micro-Imaging System (VisualSonics Inc, Toronto, Ontario, Canada). Mice were first shaved on the abdomen and then a gel was applied to the skin over the spleen. A transducer with central frequency at 30 MHz transducer was used for imaging the spleen. Cine loops of ultrasound image were recorded digitally and reviewed. An image frame (or in the case of a very large spleen, several image frames) showing the spleen was selected for analysis of splenic size (transverse and sagital measurements).
Statistical Analysis
Error bars represent experiments performed at least in triplicate to obtain standard deviations and to calculate the standard error of the mean (SEM). Student's t-test was used where appropriate to determine statistical significance, p ≤ 0.05.
RESULTS
DNA Content, Immunophenotype, and Spleen Size as a Function of Age
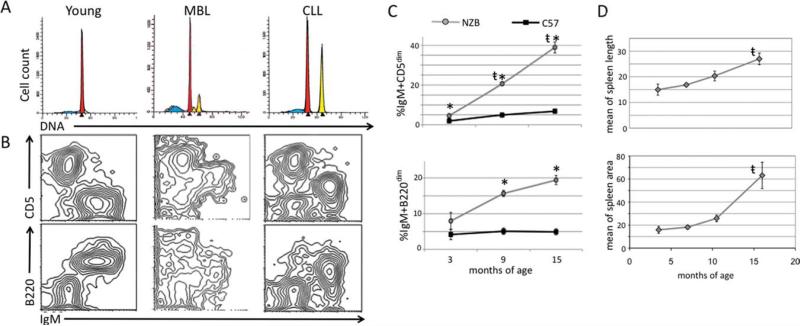
Spleens were isolated from NZB and C57Bl/6 mice at ages 3 months, 9 months, and 15 months (three mice per group), made into single cell suspensions, and analyzed for DNA content and the presence of malignant B-1 cells. Flow cytometric analysis revealed that NZB mice develop an expansion of hyperdiploid B-1 clones with age, progressing from a pauciclonal MBL state to CLL with a dominant clone (Fig. 1A), characterized as IgM+B220dimCD5dim (Fig. 1B). Aging NZB spleens had significantly higher levels of B-1 cells when compared to the spleen cells from age-matched C57Bl/6 mice (Fig. 1C). NZB mice were imaged via ultrasound from ages 3.5 months to 16 months. Spleen length and area were measured and revealed an increase in splenomegaly in the NZB with age with a dramatic increase in area after 10 months of age (Fig. 1D).
Fig. 1.
Analysis of DNA content, immunophenotype, and spleen size with age. A, Representative analysis of DNA content from young (3 months) disease-free, MBL (9 months), and CLL (15 months) NZB spleens is shown. The first largest red peaks represent diploid G1 with the subsequent yellow peaks representing aneuploidy, (peaks shown are computer modeled). B, Representative flow cytometric analysis of surface staining of NZB splenic cells (from spleen samples above in A) for IgM, CD5, and B220. C, Lines represent the percentage of IgM+ CD5dim (top) and IgM+ B220dim (bottom) cells in the lymphoid population in NZB (gray) and C57Bl/6 (black). *Statistical significance between percentage of NZB and C57Bl/6 lymphoid populations. (t) Indicates statistical significance among aging NZB. Error bars represent the SEM for three independent stainings. D, NZB mice were imaged periodically via ultrasound from ages 3.5 months to 16 months. Spleen length (top) and area (bottom) were measured. (t) Indicates statistical significance among aging NZB.
Alteration in mir-15a/16-1 Affects miR-16 Expression
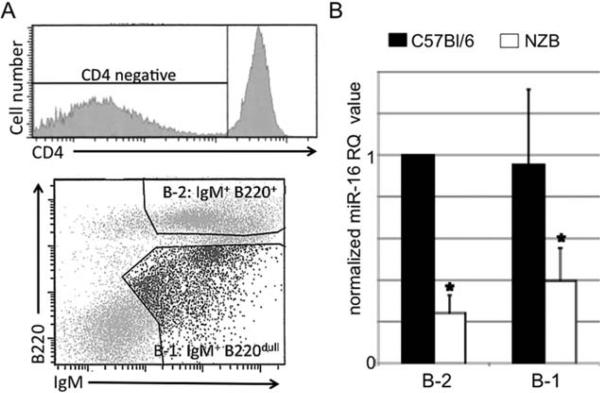
We previously found the NZB to exhibit a germline point mutation in mir-15a/16-1 on chromosome 14 (23), similar to the point mutation reported in two CLL patients (24). This mutation was found in all NZB mice sequenced (over 30 mice), present in both lymphoid and nonlymphoid tissues, as well as in NZB-derived B and T cell lines. In addition, F1 crosses with NZB mice are heterozygous for the point mutation, indicating germline transmission (data not shown). We also reported that this mutation correlated with a decrease in mature miR-16 levels in the spleen (23). Here, we analyzed sorted subpopulations containing both nonmalignant and malignant B cells to see if the miR-16 decrease was specific to B-1 cells. Spleen cells obtained from both NZB and C57Bl/6 mice (10-12 months of age) were sorted into conventional B-2 cells (CD4− , IgM+, B220+) or B-1 cells (CD4− , IgM+ B220dim) (Fig. 2A). Real time PCR revealed both NZB conventional B-2 cells and B-1 cells (containing the malignant B clone) to have less than half the amount of miR-16 expression relative to the B-2 and B-1 cell populations from age-matched C57Bl/6 spleens (Fig. 2B), indicating that all B cells from NZB mice are affected by the point mutation in the mir-15a/16-1 locus.
Fig. 2.
miR-16 expression. A, A representative sort from an individual NZB spleen is shown. The initial population of CD4− Cells (top) stained for IgM and B220 (CD45R) (bottom) were sorted into conventional B-2 (IgM+, B220+) and B-1 cells (IgM+, B220dim). C, miR-16 expression analysis in each sorted B cell population is reported as the normalized RQ 2−ΔΔCT value relative to the expression in the C57Bl/6 B-2 population (C57Bl/6 B-2 1). (Black columns are C57Bl/6 and white are NZB values). Error bars represent the SEM for three independent sorts. *Represents statistical significance between miR-16 levels of both NZB B-1 and B-2 cells compared to C57 B-1 cells, P < 0.05).
Transfected miR-16 Enhanced Drug Sensitivity
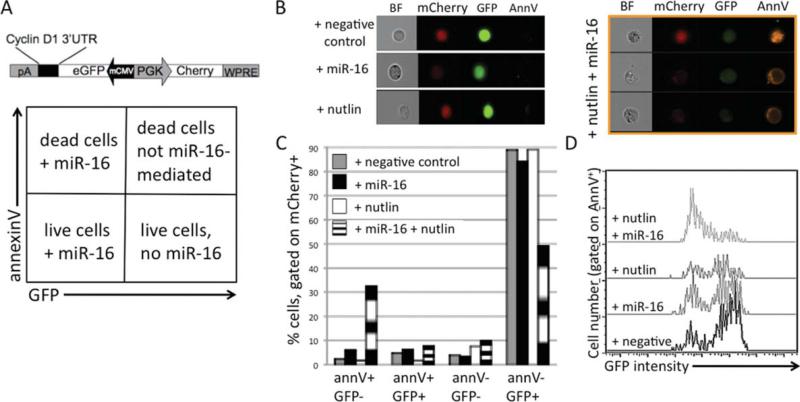
We have previously found miR-16 restoration to cause a G1 arrest and a decrease in cyclin D1 protein levels in our NZB-derived malignant B-1 cell line, LNC, by direct interaction of the miR-16 with the cyclin D1 3′UTR target (27). We have also found miR-16 and nutlin to synergistically augment apoptosis in LNC (27). Here, we transduced LNC with the bi-directional lentiviral vector that expresses both mCherry and GFP, and contains the miR-16 target sites of the cyclin D1 3′UTR downstream of the GFP expression cassette (Bd.LV.GFP.D1, Fig. 3A) as previously described (27). To visualize the apoptotic effects of miR-16 and nutlin, an MDM2 antagonist and p53 activator (28), the BD.LV.GFP.D1 transduced LNC were then transfected with either negative control mimic or miR-16 mimic (Dharmacon) and incubated 48h in the presence of 5 μM nutlin. To detect the efficiency of miR-16 transfection and induction of apoptosis, transduced cells were stained with annexin V PE and analyzed using the Amnis ImageStream, gating on mCherry-positive transduced cells. The addition of negative control mimic or nutlin alone had no effect on GFP or mCherry (internal control) expression, as expected. Cells staining positive for annexin V, but GFP−/dim, died from miR-16 transfection. GFP+ cells also staining positive for annexin V, died via a non-miR-16 mediated mechanism, perhaps from electroporation or from nutlin. Cells that were negative for GFP and annexin V were most likely live cells that were transfected with miR-16. Cells bright for GFP, but negative for annexin V were live cells that were not successfully transfected with miR-16 (Fig. 3A). The addition of miR-16 resulted in a decrease in GFP expression signal (Fig. 3B), indicating successful miR-16 mimic activity with direct interaction with the downstream miR-16 target sites of the cyclin 3′UTR. Transfection with negative control mimic or miR-16 or treatment with nutlin alone showed little apoptotic effect on transduced cells, which is indicated in the Amnis images as annexin V− (Fig. 3B). However, induction of apoptosis was seen upon addition of both miR-16 and nutlin to the transduced cells, as indicated by the Amnis images (Fig. 3B). A greater percentage of mCherry+ GFP− transduced cells treated with both miR-16 and nutlin were annexin V positive as compared with the transduced cells with negative control, miR-16, or nutlin alone (Fig. 3C). The population treated with both nutlin and miR-16 had the lowest percentage of GFP+ live cells (annexin V− , Fig. 3C). mCherry+ cells were then gated on annexin V+ and analyzed for GFP intensity. Annexin V+ cells treated with both nutlin and miR-16 were predominantly dim for GFP, indicating that apoptosis was enhanced when both the chemotherapeutic agent and the microRNA were added together (Fig. 3D).
Fig. 3.
Transfected miR-16 enhances drug sensitivity to nutlin. A, Schematic diagram of bidirectional lentiviral vector used, Bd.LV.GFP.D1 (top). Diagram describing the population present in each quadrant (bottom). B, Single representative flow cytometric Amnis images of bright field (BF), mCherry, GFP, and annexin V (AnnV) expression of Bd.LV.GFP.D1 transduced cells transfected with negative control or miR-16 or treated with nutlin alone (left panel). Multiple Amnis images of Bd.LV.GFP.D1 transduced cells transfected with miR-16 and treated with nutlin (right panel, three cells shown). C, Cells were gated on mCherry-positive transduced cells and analyzed for annexinV and GFP expression. Columns are % of cells in the four quadrants in A. Gray = transduced cells transfected with negative control mimic, black miR-16, white nutlin, black and white striped = both nutlin and miR-16. D, Cells were gated on mCherry+ and annexinV+ and analyzed for GFP intensity.
Identification of an NZB SP
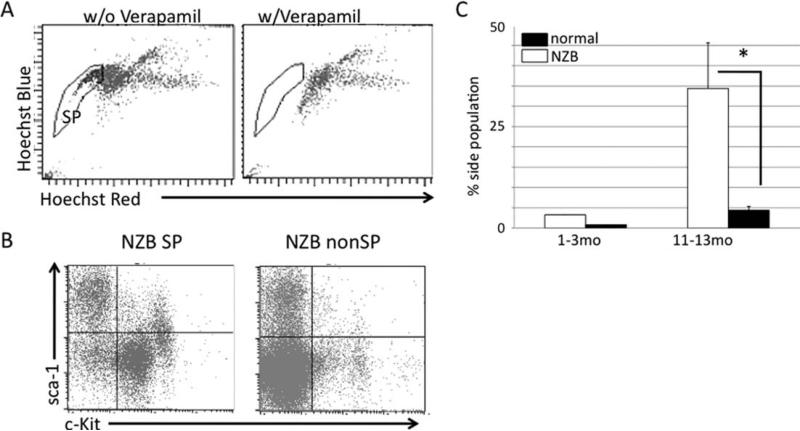
A population of cells responsible for the maintenance and progression of the NZB malignant B-1 cells was sought. Spleens were isolated from young (1–3 months old) and old (11–13 months old) NZB mice and age-matched normal strain (C57Bl/6 or DBA/2) mice. Samples were then analyzed for the presence of a side population (SP) based on their ability to exclude the Hoechst 33342 (Fig. 4A), gating on the PI negative live cell population. The NZB SP was shown to have a c-kit+ sca-1+ population that is lacking in the non-SP. The non-SP cells were mostly double negative for c-kit and sca-1 (Fig. 4B). The SP in the NZB spleen increases with age, with the aged NZB having a significantly higher percentage of SP cells when compared with age-matched normal strain mice (Fig. 4C). Spleens isolated from three NZB mice were also sorted into SP or non-SP and RNA was analyzed for the expression of miR-16, miR-181a, and miR-155 via real time PCR. miR-155 is found to be elevated in patients with B-cell lymphoma (36–38) as well as in the NZB (data not shown). miR-181a is increased in differentiated B cells (39). While no difference was observed in miR-16 levels between SP and non-SP, miR-181a was much lower in SP cells and miR-155 was higher in SP cells when compared to the non-SP (data not shown).
Fig. 4.
Identification of NZB SP. A, Representative flow analysis of the presence of SP (left panel) that loses its ability to exclude Hoechst 33342 in the presence of Verapamil (right panel). B, Representative flow cytometric analysis of NZB SP and non-SP cells for expression of sca-1 and c-kit. C, Percentage of SP cells in the spleen. Black columns are normal (C57Bl/6 or DBA/2) strain and white is NZB. Error bars represent SEM, three mice per group. *Significant difference in % SP between old NZB and old normal strain mice (P < 0.05).
CFU-Pre-B Cell Assay on Sorted SP and Non-SP
SP cells negative for B cell lineage markers were then assayed for their ability to generate B-1 cells in vitro. Splenic cells from a 17 month-old NZB and from three pooled 8-10 month-old C57Bl/6 mice were gated on 7AAD−/IgM− and sorted into the following populations: SP B220−, SP B220+, non-SP B220−, and non-SP B220+, (Fig. 5A). Each of the four populations was then cultured in Methocult methylcellulose medium for mouse pre-B cell colonies. After a 7-day incubation, colonies were counted and analyzed on the basis of size. The SP B220− population had the largest colonies consisting of over 50 cells for both NZB and C57Bl/6 (data not shown). After 7 days of culture of the SP B220− population in mouse pre-B cell media, cells were then harvested and analyzed for expression of IgM, CD5, and B220. The majority of the Methocult-cultured C57Bl/6 subpopulations were IgM− CD5− B220− (Fig. 5B). In contrast, the majority of NZB SP B220− Cultured cells were able to differentiate into pre-B-1 cells which expressed B220 and CD5, but did not express IgM (Fig. 5B). These results indicate that the SP population is not only increased in NZB spleens, but also, that the NZB SP cells contain a B-1 stem cell.
Fig. 5.
CFU-pre-B cell assay on sorted SP and non-SP. A, Flow cytometry cell sorting scheme for spleen cells which were 7AAD− and gated on IgM− and sorted into SP and non-SP, and then further into SP B220− population (17-month-old NZB shown as an example). B, Sorted cells were grown in Methocult pre-B cell media for 7 days and cells were harvested. Representative flow cytometric contour plots for NZB SP cells (top rows) and C57Bl/6 SP cells (bottom rows) harvested from 7 days Methocult culture. Percentage of cells in each quadrant is indicated.
In Vivo SP and Non-SP Transfer
Since our data suggested that the NZB SP contained a population of B-1 progenitor cells, we decided to test the ability of these cells to transfer malignancy. To determine if SP cells can better transfer disease than non-SP cells, 4-week-old NZB mice (three per treatment group) were injected with 2 × 106 sorted SP or non-SP cells from the hyperdiploid spleen of a 13-month-old NZB donor with malignant B-1 cell (IgM+ CD5dim) expansion (Fig. 6A). Before injection, the sorted SP and non-SP populations were analyzed for DNA content. The SP revealed only a single G1 diploid peak, whereas the non-SP revealed multiple G1 peaks, indicative of hyperdiploidy (Fig. 6B). On the basis of DNA content, we concluded that the NZB SP did not contain the hyperdiploid clone, whereas the non-SP did. The donor NZB SP contained no mature CD19+ B cells, but rather, immature pre-B cells, expressing B220, whereas the non-SP contained both CD19+ B cells, B220 bright and dim populations (Fig. 6B). NZB recipients were evaluated 5.5 months postinjection for splenomegaly, hyperdiploidy, and presence of B-1 cells. Mice injected with non-SP cells exhibited splenomegaly with the spleen/body weight ratio more than doubling when compared to SP recipient mice (Fig. 6C). The spleens of SP recipients had a lower percentage of CD19+ CD5− B-2 cells of total B cells, while having more CD19+ CD5dim B-1 cells when compared with the non-SP recipients (Fig. 6D). However, whether the mouse was injected with either SP or non-SP, the total B cell population contained more B-2 cells. The peritoneal wash cells (PWC) of SP recipients had a lower percentage of B-2 cells of total B cells, and a higher level of B-1 cells when compared with non-SP recipients (Fig. 6D). Upon histological analysis, it was noted that the spleens of mice either injected with SP or non-SP had large germinal centers and an increase in Mott cells (indicated by arrow) as compared with the control mouse. Spleens of mice injected with SP had more white pulp when compared with control or non-SP mice (Fig. 6E). The liver of mice injected with either SP or non-SP contained increased foci of lymphocytes (as indicated by arrow) when compared with the control. Foci appeared larger in mice injected with non-SP (Fig. 6E). Lymphocyte infiltration in the kidneys was noted when mice were injected with SP or non-SP, with the kidney of one non-SP mouse having significantly more glomeruli with infiltrates (data not shown). Only one mouse, an SP recipient, developed 13.8% aneuploidy (data not shown).
Fig. 6.
In vivo transfer of NZB SP and non-SP. A, Top row: hypotonic PI analysis of DNA content of spleen (left panel) and PWC (right panel) of 13mo NZB donor. Peaks represent cells in G1, with spleen having a diploid G1 peak (red) and a dominant hyperdiploid peak (yellow). Bottom row: Flow cytometric analysis of 13mo NZB donor spleen (left) and PWC (right) stained with antibodies against IgM and CD5. Percentage of each cell type of lymphoid gate is indicated in the quadrant. B, The spleen from the 13 month NZB donor was sorted into SP and non-SP. Right panel: DNA analysis of sorted non-SP (top) and SP (bottom) from donor NZB showing G1 hyperdiploidy present only in the non-SP population. SP and non-SP cells from the donor NZB were also analyzed via flow cytometry for expression of CD19, B220, and sca-1. C, Ratio of spleen to body weight at time of sacrifice. White column = ratio of NZB injected with SP and black = ratio of NZB injected with non-SP. Error bars represent SEM, 3 mice/group. D, Top row: Representative flow cytometric analysis of spleens and PWC from NZB recipients injected with SP or non-SP sorted cells. Bottom row: B-2 (CD19+ CD5− ) and B-1 (CD19+ CD5dim) cells are shown as a percentage of total B cells from spleen (left) and PWC (right) of NZB recipients injected with SP (white bars) or non-SP (black bars). Error bars are SEM, three mice per group. E, Representative histological analysis of spleen (top row) and liver (bottom row) from control, SP, and non-SP recipient mice stained with hematoxylin and eosin, (×40 for spleen, ×10 for the control liver, and ×20 for experimental liver with foci). Arrows in spleen indicate presence of Mott cells, and arrows in liver sections indicate lymphocyte foci.
DISCUSSION
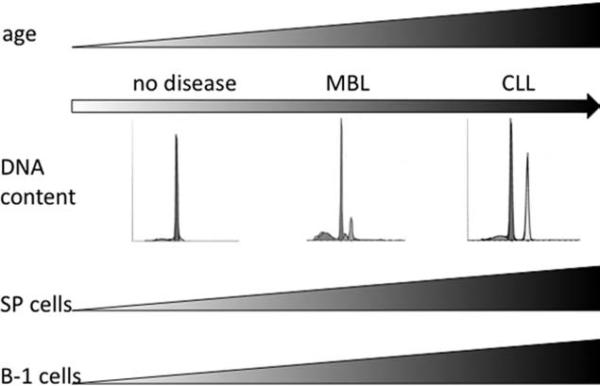
In this report, we find that the NZB spleen exhibits a clonal expansion of hyperdiploid IgM+ CD5dim B220dim B-1 cells with age. Initially, the NZB develops a B-cell lymphoproliferative disorder similar to MBL in humans, at which multiple hyperdiploid B-1 clones are present. As the disease progresses, a single malignant B-1 cell clone dominates, developing into CLL (Fig. 7). In humans, MBL is as an indolent hematologic disease that may never fully develop into CLL. Although the NZB mice cannot be used as a model for stable MBL because of their inevitable progression to CLL, they can be used to study MBL as a precursor to CLL and the age-associated development and progression from MBL to CLL.
Fig. 7.
Model for NZB disease progression from MBL to CLL. Young NZB exhibit no detectable disease: no hyperdiploidy and low percentages of SP and IgM+ CD5+ B-1 cells. As NZB age, they begin to manifest an MBL-like disease characterized by an expansion of multiple hyperdiploid B-1 clones and an increase in the percentage of SP cells. MBL disease progresses to CLL in old NZB. These mice have a single dominant malignant B-1 cell clone and a large percentage of SP and B-1 cells.
The NZB exhibits a T→A point mutation six bases downstream from pre-mir-16-1 on mouse chromosome 14 (23), similar to the C→T point mutation reported in two human CLL patients on human chromosome 13 (24). In the majority of human CLL patients, the deletion of the 13q14 region results in a loss of the mir-15a/16-1 locus. Both types of genetic alterations, deletion or mutation, result in a decrease in mature miR-15a and miR-16.
Since the NZB mutation is germline, all NZB cells have the point mutation in mir-15a/16-1 rather than a chromosomal deletion (23). Due to the presence of a homolog containing nearly identical miRNAs, mir-15b/16-2, in both humans and mice, the expression of mature miR-16 can be derived from either homolog and is tissue and cell dependent. The kidney, for instance, expresses miR-16 derived from the mir-15b/16-2 locus. In contrast, B cells utilize the mir-15a/16-1 locus. While NZB B-2 and B-1 cell populations have decreased levels of mature miR-15a/16 when compared with normal strain mice, only the B-1 cells develop clones that contribute to MBL development. Decreased miR-15a/16 alone may not be sufficient for MBL progression to CLL; however, decreased miR-15a/16 is sufficient for B cell clonal development, as seen in both human MBL and NZB MBL.
We propose that this decrease in mature miR-16 is a result of improper processing caused by the point mutation in the mir-16-1 flanking region. The single-stranded RNA stem loop flanking regions are important for Drosha processing and this point mutation may shift the recognition site for Drosha cleavage (40–42). Our data supports a role for the miR-16 defect in cell cycle progression of the malignant cells due to its direct interaction with the cyclin D1 3′UTR and resulting G1 arrest when exogenously delivered to the NZB-derived malignant B-1 cell line (27). This miRNA defect may also play a role in the chemoresistance of the NZB malignant B-1 cells. Here, with the use of a lentiviral sensor system, we show that only the NZB cells that are successfully transfected with miR-16 are sensitized to the apoptotic effects of a chemotherapeutic agent, nutlin. miR-16 synergizes with nutlin to enhance apoptosis of the NZB malignant B-1 cells, as indicated by the mCherry+ annexinV+ GFPdim cells.
We also show the NZB to have an increase in “side population” cells that possess the ability to exclude Hoechst 33342 dye when compared with non-NZB spleen sources. The NZB contain a large SP that dramatically increases with age (Fig. 7), yet does not contain the hyperdiploid B-1 clone. Rather, the NZB SP contains a population of B-1 progenitors. The NZB sorted SP cells that were B cell lineage negative were able to differentiate into B lineage cells in the presence of B cell growth factors. After 7 days of culture, the NZB SP cells that were initially B220−/IgM− differentiated into greater than 90% B220+ cells, which were also CD5+. In contrast, very few C57Bl/6 SP cells that were initially B220 negative were able to differentiate into pre-B cells, and more importantly, did not express CD5. This data suggests that the NZB SP cells, when stimulated to undergo B lymphopoiesis in vitro, predominantly generate pre-B-1 cells. Previous reports have shown that as NZB age, the bone marrow of NZB contain B220− stem cells that have reduced kinetics in differentiating into mature B cells (43). Contrary to this finding, when cultured in Methocult, the NZB SP B220− did not develop pre-B cells at a slower rate than the C57 SP B220− population. Although our in vivo transfer data show a slight increase in B-1 development in the NZB SP recipients, we did not see an increased severity or early development of the CLL disease. Recipients of the NZB non-SP exhibited splenomegaly, yet this does not appear to be solely a result of B-1 expansion. We speculate that the increase in SP with age, which also correlates with age-associated splenomegaly and AIHA, may be the result of both B cell expansion and extramedullary hematopoiesis. We propose that the SP cells are an indication of this imbalance between white and red blood cells and B cell hyperactivity. In addition, in order to transfer malignancy, other populations of cells, such as T-regulatory cells (Tregs), may also be needed. Tregs have immunosuppressive effects, and transfer of Tregs would suppress the recipient's immune system from attacking the malignant cells, favoring tumor development and growth (44,45). miR-16 does not seem to be correlated with this increase in SP due to the fact that sorted NZB SP and non-SP subgroups exhibit no difference in miR-16 levels. However, NZB non-SP cells contain much higher levels of miR-181a, consistent with an increase in this population of differentiated B cells (39). In contrast, higher levels of miR-155 were found in the SP population, and miR-155 is also found to be elevated in hematopoietic stem cells and the bone marrow of patients with B-cell lymphoma (36–38).
Although, the side population has been linked to a population of leukemia-initiating cells in patients with acute myelogenous leukemia (46), no “cancer stem cell” has yet successfully been isolated in CLL. However, Foster et al. has shown the loss of a side population to precede the loss of malignant CLL cells during the course of chemotherapy (47), with the SP perhaps playing a supportive role in chemo-resistance. The NZB SP cells did not seem to contain cancer stem cells because sorted SP cells did not contain the malignant B-1 clone nor reconstitute CLL disease when injected into young NZB mice, although one recipient of NZB SP cells did develop hyperdiploid B-1 cells, suggesting that the malignant clone did develop in this recipient. Although the NZB SP may not contain actual cancer stem cells, we propose that they contain a population of cells that foster the growth and expansion of malignant B-1 cells, aiding in the age-associated disease progression from MBL to CLL. A similar population of cells referred to as fat-associated lymphoid clusters (FALC) were recently identified in the peritoneal cavity of mice. These cells expressed c-kit and sca-1 but no lineage markers, and produced IL-5, which is important in B-1 cell maintenance and self-renewal (48). Further studies need to be performed to define a role for the SP cells in disease development and to determine if they can transfer malignancy.
In this report, we support the use of the NZB mouse as a model for studying the development and progression of CLL. At present, there is no well-defined mouse model for MBL; however, the NZB may be utilized in studying MBL-like disease and progression. Similar to CLL, the NZB develop an age-associated expansion of hyperdiploid malignant CD5+ B-1 cells, progressing from a pauciclonal MBL-like state to monoclonal CLL. Alterations in the primary mir-15a/16-1 locus of DLEU2 have been linked to the development of CLL in the NZB murine model (23), and reduced levels of mature miR-16 in NZB spleen and B cell subpopulations have been reported. 13q14 deletions, containing mir-15a/16-1, were also reported in patients with MBL (16), suggesting that this chromosomal aberration and resultant loss of mir-15a/16-1 is an early event in the development of MBL, as well as being frequently present in CLL. The underlying deficiency in mir-15a/16-1 could lead to increased proliferation since our research and other reports have shown that cyclin D1 is targeted by miR-15a/16 (27,49,50). Decreased levels of miR-16 are also associated with a lack of sensitivity to drugs that target proliferation and apoptotic pathways (27,49). It is possible that in the precursor MBL state, the loss of miR-15a/16 may contribute to a failure to regulate the clonal development. Additional triggers may drive these abnormal MBL clones, harboring the mir-15a/16-1 deletion to progress to CLL.
ACKNOWLEDGMENTS
The authors thank Brian D. Brown, Alessia Baccarini, Sukwinder Singh, and Howard S. Mostowski. This research was in part funded by NIH NCI R01CA129826 granted to E. Raveche and NJCCR pre-doctoral fellowship 09-1255-CCR-E0 awarded to E. Salerno.
LITERATURE CITED
- 1.Ishibe N, Sgambati MT, Fontaine L, Goldin LR, Jain N, Weissman N, Marti GE, Caporaso NE. Clinical characteristics of familial B-CLL in the National Cancer Institute Familial Registry. Leuk Lymphoma. 2001;42:99–108. doi: 10.3109/10428190109097681. [DOI] [PubMed] [Google Scholar]
- 2.Caporaso N, Marti GE, Goldin L. Perspectives on familial chronic lymphocytic leukemia: Genes and the environment. Semin Hematol. 2004;41:201–206. doi: 10.1053/j.seminhematol.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Sellick GS, Catovsky D, Houlston RS. Familial chronic lymphocytic leukemia. Semin Oncol. 2006;33:195–201. doi: 10.1053/j.seminoncol.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Chiorazzi N, Rai K, Ferrarini M. Mechanisms of disease: Chronic lymphocytic leukemia. New Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 5.Caligaris-Cappio F, Ghia P. The normal counterpart to the chronic lymphocytic leukemia B cell. Best Pract Res Clin Haematol. 2007;20:385–397. doi: 10.1016/j.beha.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 8.Bullrich F, Fujii H, Calin G, Mabuchi H, Negrini M, Pekarsky Y, Rassenti L, Alder H, Reed JC, Keating MJ, Kipps TJ, Croce CM. Characterization of the 13q14 tumor suppressor locus in CLL: Identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res. 2001;61:6640–6648. [PubMed] [Google Scholar]
- 9.Liu Y, Corcoran M, Rasool O, Ivanova G, Ibbotson R, Grander D, Iyengar A, Baranova A, Kashuba V, Merup M. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromo-some 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene. 1997;15:2463–2473. doi: 10.1038/sj.onc.1201643. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicoloso MS, Kipps TJ, Croce CM, Calin GA. MicroRNAs in the pathogeny of chronic lymphocytic leukaemia. Br J Haematol. 2007;139:709–716. doi: 10.1111/j.1365-2141.2007.06868.x. [DOI] [PubMed] [Google Scholar]
- 12.Rawstron AC, Yuille MR, Fuller J, Cullen M, Kennedy B, Richards SJ, Jack AS, Matutes E, Catovsky D, Hillmen P, Houlston RS. Inherited predisposition to CLL is detectable as subclinical monoclonal B-lymphocyte expansion. Blood. 2002;100:2289–2290. doi: 10.1182/blood-2002-03-0892. [DOI] [PubMed] [Google Scholar]
- 13.Marti G, Abbasi F, Raveche E, Rawstron AC, Ghia P, Aurran T, Caporaso N, Shim YK, Vogt RF. Overview of monoclonal B-cell lymphocytosis. Br J Haematol. 2007;139:701–708. doi: 10.1111/j.1365-2141.2007.06865.x. [DOI] [PubMed] [Google Scholar]
- 14.Marti GE, Carter P, Abbasi F, Washington GC, Jain N, Zenger VE, Ishibe N, Goldin L, Fontaine L, Weissman N, Sgambati M, Fauget G, Bertin P, Vogt RF, Jr, Slade B, Noguchi PD, Stetler-Stevenson MA, Caporaso N. B-cell monoclonal lymphocytosis and B-cell abnormalities in the setting of familial B-cell chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2003;52B:1–12. doi: 10.1002/cyto.b.10013. [DOI] [PubMed] [Google Scholar]
- 15.Shim YK, Middleton DC, Caporaso NE, Rachel JM, Landgren O, Abbasi F, Raveche ES, Rawstron AC, Orfao A, Marti GE, Vogt RF. Prevalence of monoclonal B-cell lymphocytosis: a systematic review. Cytometry B Clin Cytom. 2010;78B(Suppl 1):S10–S18. doi: 10.1002/cyto.b.20538. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanasa MC, Allgood SD, Volkheimer AD, Gockerman JP, Whitesides JF, Goodman BK, Moore JO, Weinberg JB, Levesque MC. Single-cell analysis reveals oligoclonality among ‘low-count’ monoclonal B-cell lymphocytosis. Leukemia. 2010;24:133–140. doi: 10.1038/leu.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips JA, Mehta K, Fernandez C, Raveche ES. The NZB mouse as a model for chronic lymphocytic leukemia. Cancer Res. 1992;52:437–443. [PubMed] [Google Scholar]
- 18.Scaglione BJ, Salerno E, Balan M, Coffman F, Landgraf P, Abbasi F, Kotenko S, Marti GE, Raveche ES. Murine models of chronic lymphocytic leukaemia: Role of microRNA-16 in the New Zealand Black mouse model. Br J Haematol. 2007;139:645–657. doi: 10.1111/j.1365-2141.2007.06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang AM, Phillips JA, Lin T, Raveche ES. Altered CD45 expression in malignant B-1 cells. Cell Immunol. 1996;169:196–207. doi: 10.1006/cimm.1996.0110. [DOI] [PubMed] [Google Scholar]
- 20.Ramachandra S, Metcalf RA, Fredrickson T, Marti GE, Raveche E. Requirement for increased IL-10 in the development of B-1 lymphoproliferative disease in a murine model of CLL. J Clin Invest. 1996;98:1788–1793. doi: 10.1172/JCI118978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theofilopoulos AN. Genetics of systemic autoimmunity. J Autoimmun. 1996;9:207–210. doi: 10.1006/jaut.1996.0025. [DOI] [PubMed] [Google Scholar]
- 22.Ghia P, Scielzo C, Frenquelli M, Muzio M, Caligaris-Cappio F. From normal to clonal B cells: Chronic lymphocytic leukemia (CLL) at the crossroad between neoplasia and autoimmunity. Autoimmun Rev. 2007;7:127–131. doi: 10.1016/j.autrev.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, Fredrickson T, Landgraf P, Ramachandra S, Huppi K, Toro JR, Zenger VE, Matcalf RA, Marti GE. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassnti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 25.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 27.Salerno E, Scaglione BJ, Coffman FD, Brown BD, Baccarini A, Fernandes H, Marti G, Raveche ES. Correcting miR-15a/16 genetic defect in New Zealand Black mouse model of CLL enhances drug sensitivity. Mol Cancer Ther. 2009;8:2684–2692. doi: 10.1158/1535-7163.MCT-09-0127. [DOI] [PubMed] [Google Scholar]
- 28.Coll-Mulet L, Iglesias-Serret D, Santidrian AF, Cosialls AM, de Frias M, Castano E, Campas C, Barragan M, de Sevilla AF, Domingo A, Vassilev LT, Pons G, Gil J. MDM2 antagonists activate p53 and synergize with genotoxic drugs in B-cell chronic lymphocytic leukemia cells. Blood. 2006;107:4109–4114. doi: 10.1182/blood-2005-08-3273. [DOI] [PubMed] [Google Scholar]
- 29.He H, Chen L, Zhai M, Chen JZ. Genistein down-regulates the constitutive activation of nuclear factor-kappaB in human multiple myeloma cells, leading to suppression of proliferation and induction of apoptosis. Phytother Res. 2009;23:868–873. doi: 10.1002/ptr.2715. [DOI] [PubMed] [Google Scholar]
- 30.Hadnagy A, Gaboury L, Beaulieu R, Balicki D. SP analysis may be used to identify cancer stem cell populations. Exp Cell Res. 2006;312:3701–3710. doi: 10.1016/j.yexcr.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 31.Challen GA, Boles N, Lin KK, Goodell MA. Mouse hematopoietic stem cell identification and analysis. Cytometry Part A J Int Anal Cytol. 2009;75A:14–24. doi: 10.1002/cyto.a.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, Nakauchi H, Taniguchi H. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 34.Tarnok A, Ulrich H, Bocsi J. Phenotypes of stem cells from diverse origin. Cytometry Part A. 2010;77A:6–10. doi: 10.1002/cyto.a.20844. [DOI] [PubMed] [Google Scholar]
- 35.Peng B, Sherr DH, Mahboudi F, Hardin J, Wu YH, Sharer L, Raveche ES. A cultured malignant B-1 line serves as a model for Richter's syndrome. J Immunol. 1994;153:1869–1880. [PubMed] [Google Scholar]
- 36.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, Maggio R, Peragrin N, Santangelo S, Mauro FR, Landgraf P, Tuschl T, Weir DB, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Guarini A, Foa R, Macino G. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 38.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 39.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 40.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III. Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 42.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 43.Merchant MS, Garvy BA, Riley RL. B220- bone marrow progenitor cells from New Zealand black autoimmune mice exhibit an age-associated decline in Pre-B and B-cell generation. Blood. 1995;85:1850–1857. [PubMed] [Google Scholar]
- 44.Choileain NN, Redmond HP. Regulatory T-cells and autoimmunity. J Surg Res. 2006;130:124–135. doi: 10.1016/j.jss.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 45.Beyer M, Kochanek M, Darabi K, Popov A, Jensen M, Endl E, Knolle PA, Thomas RK, von Bergwelt-Baildon M, Debey S, Hallek M, Schultze JL. Reduced frequencies and suppressive function of CD4+CD25hi regulatory T cells in patients with chronic lymphocytic leukemia after therapy with fludarabine. Blood. 2005;106:2018–2025. doi: 10.1182/blood-2005-02-0642. [DOI] [PubMed] [Google Scholar]
- 46.Moshaver B, van Rhenen A, Kelder A, van der Pol M, Terwijn M, Bachas C, Westra AH, Ossenkoppele GJ, Zweegman S, Schuurhuis GJ. Identification of a small subpopulation of candidate leukemia-initiating cells in the side population of patients with acute myeloid leukemia. Stem Cells. 2008;26:3059–3067. doi: 10.1634/stemcells.2007-0861. [DOI] [PubMed] [Google Scholar]
- 47.Foster AE, Okur FV, Biagi E, Lu A, Dotti G, Yvon E, Savoldo B, Carrum G, Andreeff M, Goodell MA, Heslop HE, Brenner MK. Selective depletion of a minor subpopulation of B-chronic lymphocytic leukemia cells is followed by a delayed but progressive loss of bulk tumor cells and disease regression. Mol Cancer. 2009;8:106. doi: 10.1186/1476-4598-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 49.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, DeMaria R. The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 50.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]