Figure 1. rhNPY −1002 T>G is present in a conserved portion of a NPY repressor and results in altered DNA-protein interactions and decreased amygdala NPY expression.
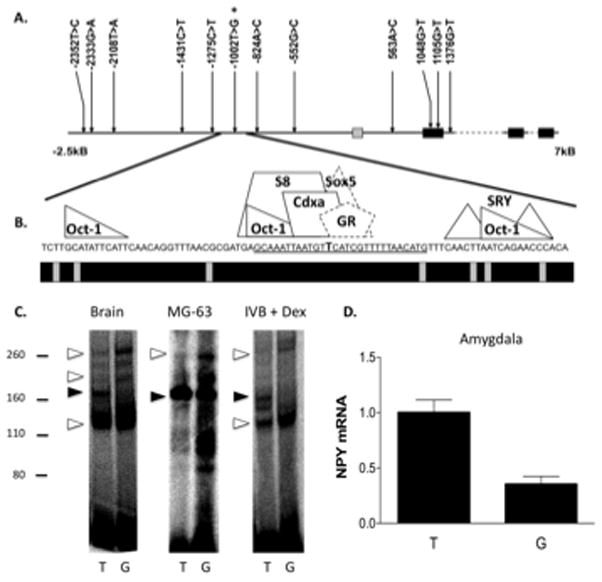
A. Schematic of the NPY gene and regulatory region and SNPs detected by sequencing of genomic DNA. B. Region 40-bp up and downstream of the −1002 T>G SNP. The precise locations of the −1002 T>G SNP (in bold) and the oligonucleotide sequence used in the gel shift assays (underlined) are indicated. Predicted sites for transcription factor binding (above) and sequence conservation among primates (below, black = conserved) are shown. Binding sites in dashed lines (Sox5 and a preferred glucocorticoid response element half site, GR) were predicted to be disrupted by the −1002 T>G SNP. C. Gel shift assay results from experiments performed using nuclear extracts from Whole Brain, osteosarcoma cells (MG-63), and glucocorticoid-treated hypothalamic cells (IVB + Dex). Relative migrations of the protein molecular weight standards are shown to the left (kDa). Open arrows indicate bands that increase with G allele probes, closed arrows indicate that which shows a relative increase with T allele probes. D. NPY mRNA expression in amygdala as a function of the −1002 T>G allele (P = 0.0008; T/T, N = 4, G carrier, N = 8). *** P < 0.001.
