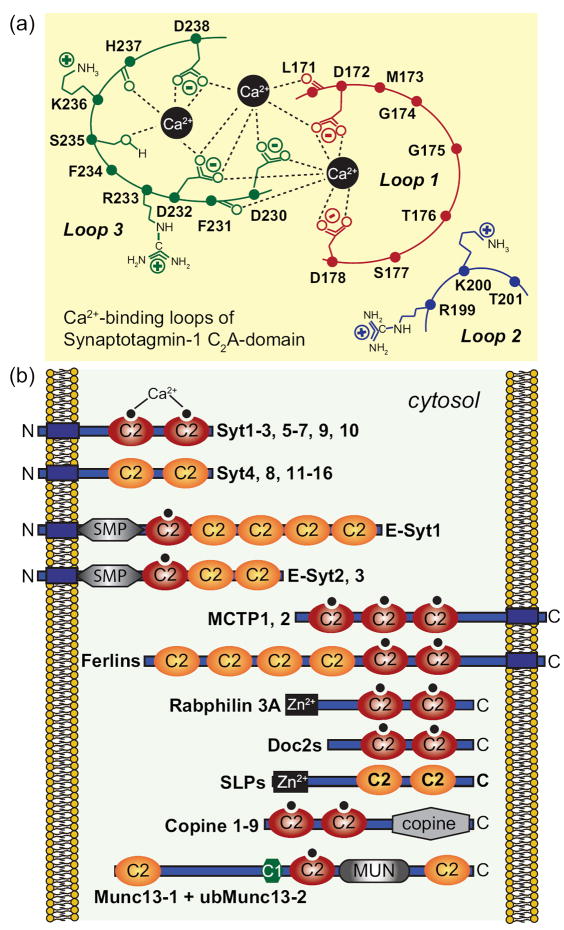
Figure 4. Structures of multiple C2-domain proteins.
A. Schematic diagram of the top Ca2+-binding loops of the Syt1 C2A-domain (modified from [41]). The C2A-domain has three top loops, of which loops 1 and 3 form three Ca2+-binding sites. Five aspartate residues, one serine residue and two backbone carbonyl groups coordinate the three bound Ca2+ ions. Residues are shown in single-letter aminio acid code and are identified by residue numbers corresponding to mouse Syt1.
B. Domain structures of proteins with multiple C2-domains that contain or lack a transmembrane domain. The mammalian genome encodes four classes of multiple C2-domain proteins containing transmembrane regions: synaptotagmins (Syt1-16; note that ‘Syt17’ does not contain a transmembrane region, but is membrane-anchored via palmitoylated cysteine residues), extended synaptotagmins (E-Syt1-3), multiple C2-domain and transmembrane proteins (MCTP1-2) and ferlins (including otoferlin, dysferlin, myoferlin and 2 other uncharacterized ferlin like proteins). Examples of soluble C2-domain containing proteins are also included. C2-domains with canonical Ca2+-binding consensus sequences are labeled as Ca2+-binding C2-domains, although Ca2+-binding has not yet been tested to all of these domains. C2-domains were assigned based on the conserved domain database of the NCBI; some proteins, especially ferlins, may have additional unpredicted C2-domains that do not precisely fit the consensus sequence, as well as alternative transcripts with fewer C2-domains.