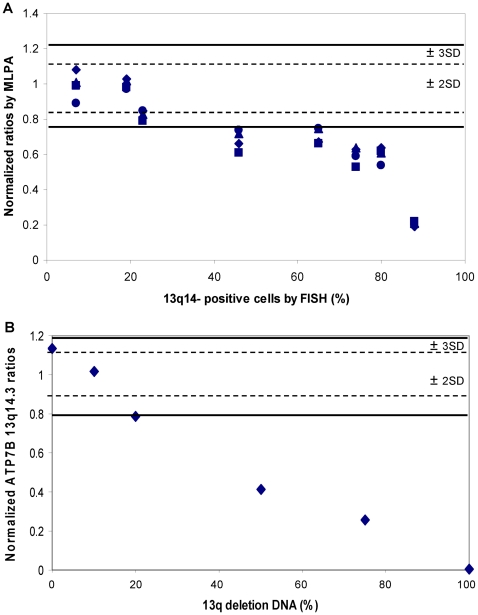
Figure 2. Correlation between MLPA and FISH analyses on 13q14- samples.
(a) Normalized ratios by MLPA were grouped by the percentage of FISH-identified leukemia clones carrying 13q14 deletion. The distribution of MLPA ratios showed that 13q14- cells must represent at least 20% (if using 2SD as cutoff) or 40% (if using 3SD as cutoff) of the total population in a sample to be called a statistically significant copy number change. (b) Mixing study using a homozygous 13q- DNA spiked into normal control DNA to create samples containing 0, 10%, 20%, 50%, 75% or 100% alterations. This study also showed a 20% detection limit on 13q deletion by MLPA.