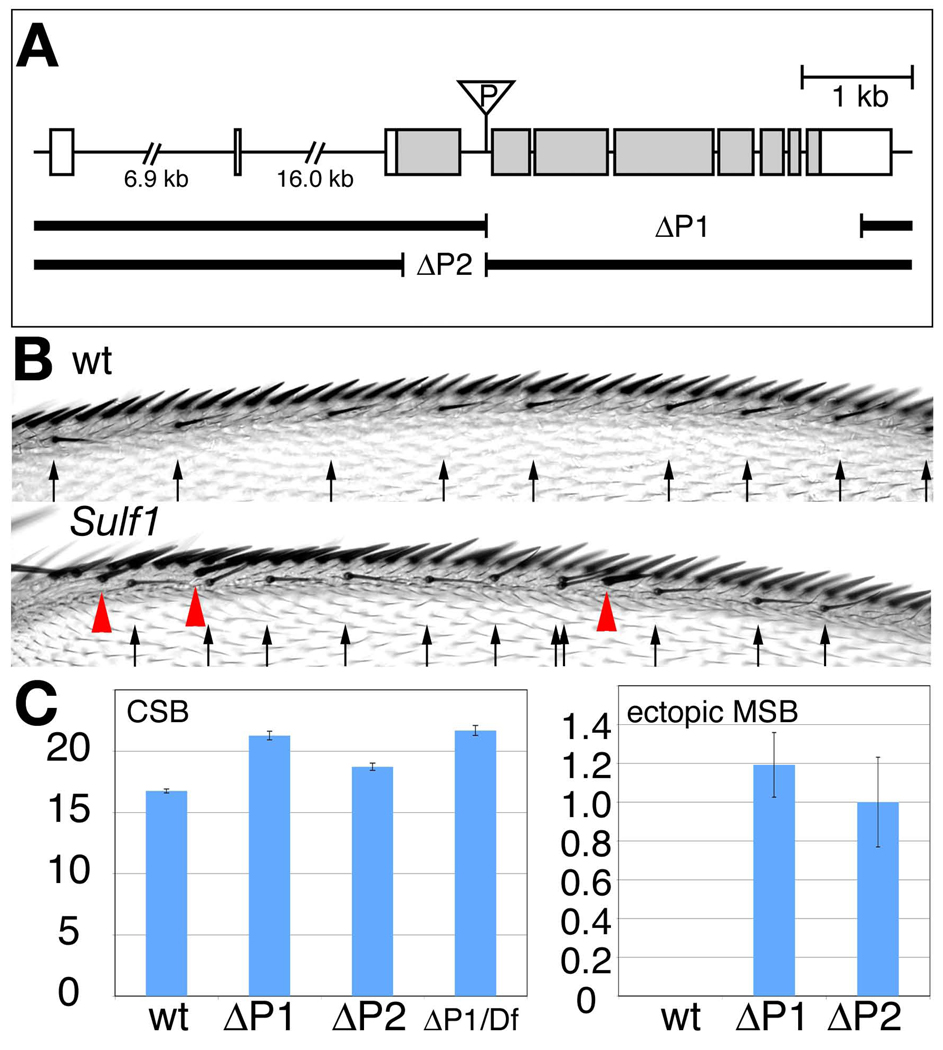
Figure 1. Phenotypes of Sulf1 mutants.
(A) Genomic organization of the Sulf1 locus. The shaded area indicates the protein coding region. Two sulf1 deletion alleles, Sulf1ΔP1 (ΔP1) and Sulf1ΔP2 (ΔP2), were generated by P-element imprecise excision. Sulf1ΔP1 lacks exon 4–10, which includes most of the protein coding region. Sulf1ΔP2 has a smaller deletion located in exon 3, which corresponds to the N-terminal domain of Sulf1 protein. This deletion causes a frame shift at amino acid residue Ser59. (B) Dorsal view of the anterior wing margin of wild-type (wt) and Sulf1 mutant adult wings. The arrows and red arrowheads indicate positions of chemosensory bristles and ectopic mechanosensory bristles, respectively. These phenotypes are consistent with an expanded region of high-level Wg signaling. (C) Bar graphs showing the number of chemosensory bristles (CSB; left) and ectopic mechanosensory bristles (MSB; right) in Sulf1 mutants. Values are shown for wild-type (wt), Sulf1ΔP1 (ΔP1), Sulf1ΔP2 (ΔP2), and Sulf1ΔP1/Df(3R)sdb26 (ΔP1/Df). Both Sulf1ΔP1 and Sulf1ΔP2 alleles had a significant increase in chemosensory (p<0.001), mechanosensory (p<0.001), and ectopic mechanosensory (p<0.001) bristles compared to wild-type. Sulf1ΔP1/Df(3R)sdb26 had a similar bristle number as Sulf1ΔP1 (p=0.2), which along with molecular data indicate that it is a null allele. Wing margin bristles were counted for more than 20 wings for each genotype. Error bars for each genotype were calculated using standard error (STDEV/SQRT(n)). P-values for the wing margin bristles were obtained using a one tailed, unpaired, Student’s t-test with equal variance in Microsoft Excel.