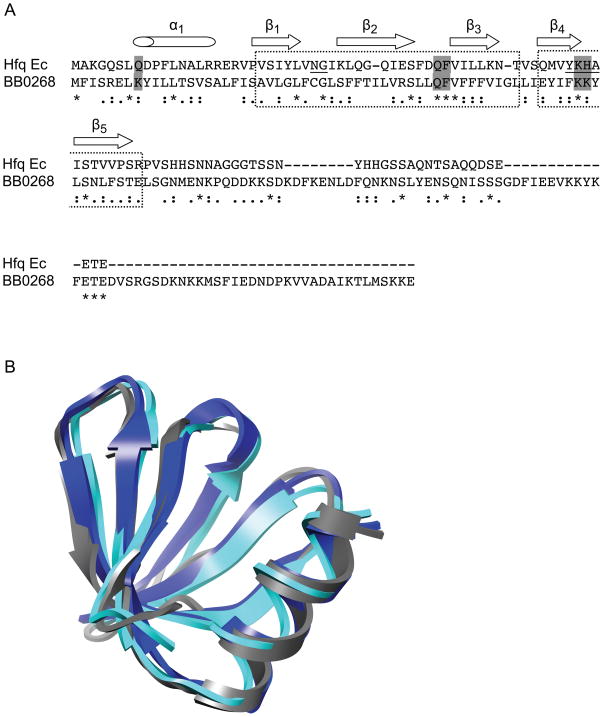
Fig. 2.
Hfq structural homology. (A) ClustalW alignment of the BB0268 conserved hypothetical protein with the E. coli (Ec) Hfq protein. Amino acids denoted by asterisks (*) are identical, colons (:) strongly similar and dots (.) weakly similar. The dashed boxes indicate the Sm1 and Sm2 motifs and the secondary structure of the E. coli Hfq is indicated above the sequence. Two of the signature motifs of Hfq, the Sm1 NG and the Sm2 F(Y)KHA, are underlined. The residues that are important in RNA binding by the S. aureus Hfq are shown in gray boxes. This figure panel is modified from Nielsen et al. (2007). (B) Modeling of Hfq structures. The structure of HfqBb (gray) was predicted using Phyre and superimposed onto the experimentally determined S. aureus Hfq (blue) and E. coli Hfq (cyan) structures using Chimera. Hfq homologs from many organisms have a variable C-terminal extension that has not been structurally characterized; therefore, we have no information on the folded structure of the C-terminal region of HfqBb and it is not shown.