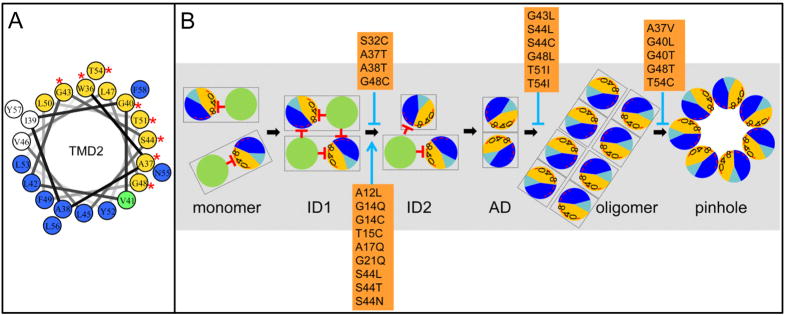
Figure 8. A. Helical wheel projection of S2168 TMD2.
Orange and dark blue colored residues are in predicted helical-helical interaction faces A and B, respectively (Pang et al., 2009). The green-colored residue (V41) is in both A and B interfaces. Red asterisks represent the position of non-lethal S2168 mutants, which are all in interface A.
B. Model for the S21 pinhole formation pathway. Same scheme as in Fig. 1C. S21 molecules are first inserted as monomers. ID1, ID2: the inactive dimers 1 and 2; AD, the activated dimer, with the homotypic helical-helical interaction of the glycine zipper containing interface A. In the oligomer state, the A:A interaction transits to A:B heterotypic helical interaction before the heptameric pinhole forms. S2168 mutants are mapped in the pathway with either accelerating the process (blue arrow), or blocking it (blue stop arrow).