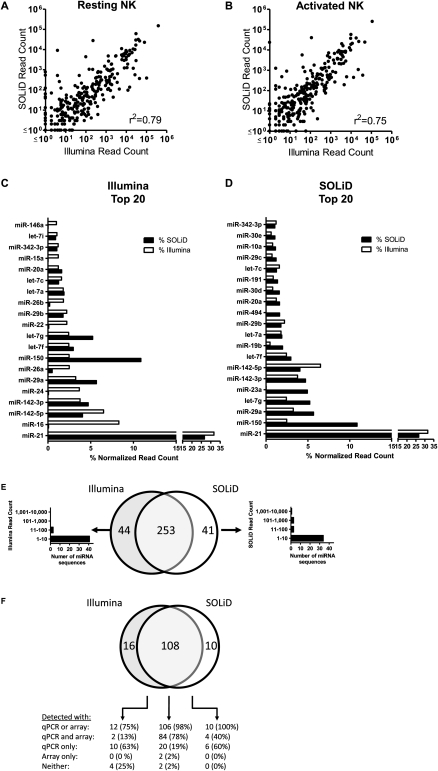
Figure 4.
Comparison of miRNA detection by Illumina GA and ABI SOLiD sequencing and hybridization-based techniques. Known miRNAs were identified in cDNA libraries created from small RNAs from resting or activated primary mouse NK cells by SHRiMP alignment to miRBase v13 hairpin precursor-miRNAs. For each miRNA detected on either platform, the dot shows the relative expression by normalized read count from Illumina (x-axis) and SOLiD (y-axis) in the resting NK cell (A) or activated NK cell (B) libraries. Overall, the correlation for detection was high (0.75). The top 20 miRNAs detected in both NK cell libraries for Illumina (C) and SOLiD (D) are shown rank ordered based on platform. (E) Venn diagram of overlapping detection in resting and activated NK cell libraries comparing Illumina and SOLiD sequencing. For miRNA detected in only one platform-data set, the normalized read count distribution is shown, highlighting that most discrepant miRNAs are expressed at relatively low levels. (F) Venn diagram showing the breakdown of overlap between Illumina GA sequencing, SOLiD sequencing, AB real-time qRT-PCR, and Agilent microarrays for 136 miRNAs with probes available for both qRT-PCR and microarrays. Detection by qRT-PCR was defined as a ΔCt (compared to MammU6) of <15, and for microarrays a normalized signal intensity of >1.0. Rare cases of major differences in the normalized read counts were detected by Illumina GA and SOLiD (Supplemental Table S4).