Figure 5.
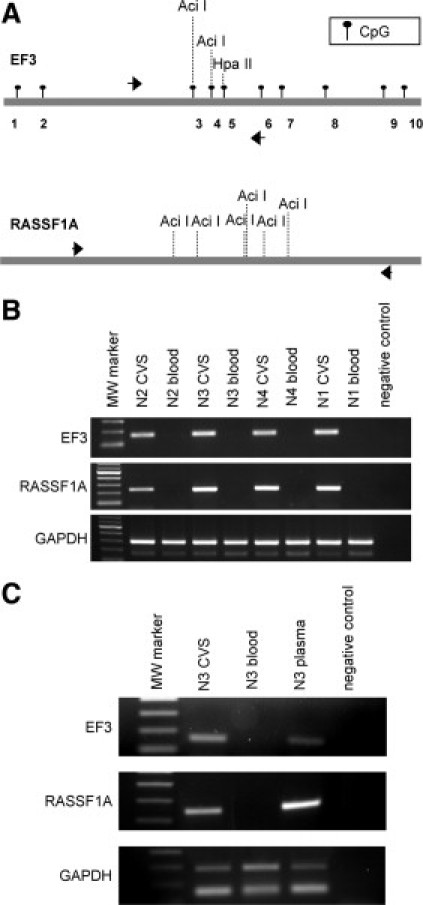
Differential detection of the EF3 and RASSF1A sequences after methylation-sensitive restriction digestion. A: Schematic representation of the EF3 and the RASSF1A sequences. The localization of CpG sites of the EF3 marker are indicated by black circles, whereas the positions of CpG sites of the RASSF1A were omitted. Black arrows represent the position of the primers used to amplify the EF3 and RASSF1A DNA after methylation-sensitive digestion. The positions of AciI and HpaII restriction sites used to discriminate between CVS and maternal blood cell DNA are reported. B: PCR of the EF3 and RASSF1A markers and the GAPDH genomic region after methylation-sensitive restriction digestion with the AciI enzyme of four CVS and maternal blood cell DNA pairs [N2 (46,XY), N3 (46,XY), N4 (46,XY), and N1 (46,XY)]. The primer pair used to amplify GAPDH provides two amplification products, the longer representing GAPDH gene (291 bp) and the shorter corresponding to pseudogene (162 bp). C: PCR of the EF3 and RASSF1A markers and the GAPDH genomic region after methylation-sensitive restriction digestion of maternal plasma, CVS, and maternal blood cell DNA of one first-trimester pregnancy [N3 (46,XY)], using the AciI and HpaII restriction enzymes. The primer pairs used are the same as illustrated in (C).
