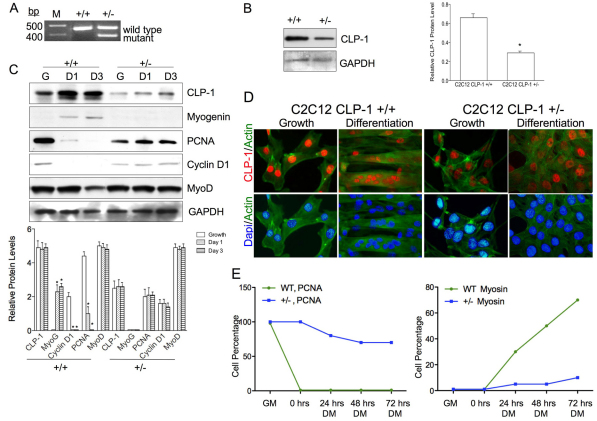
Fig. 4.
C2C12 CLP-1 +/− cells are differentiation deficient. (A) PCR using DNA from C2C12 CLP-1 +/+ cells and one CLP-1 +/− clone. Primers generate a 457 base pairs product for the CLP-1 gene and a 383 base pairs product for the mutated allele. M denotes DNA size ladder. Data are shown for one representative clone. (B) Western blot of C2C12 CLP-1 +/+ cells and C2C12 CLP-1 +/− cells in growth medium probed with anti-CLP-1 antibody. GAPDH served as a loading control. Western blot is shown for one representative clone. The analysis was performed on three independently isolated C2C12 CLP-1 +/− cell clones, and means are depicted graphically + s.e.m.; GAPDH was used for normalization (*P<0.05). (C) Western blot of lysates from C2C12 CLP-1 +/+ and +/− cell cultures in growth medium (G) and differentiation medium at 24 hours (D1) and at 72 hours (D3), probed with anti-CLP-1, anti-myogenin, anti-PCNA, anti-cyclin D1 and anti-MyoD antibodies. GAPDH served as a loading control. The analysis was performed three times, and means are depicted graphically + s.e.m.; GAPDH was used for normalization (*P<0.05, **P<0.01). (D) Immunofluorescence of C2C12 CLP-1 +/+ and C2C12 CLP-1 +/− cells in growth and differentiation medium using actin stain (green) and co-stained with anti-CLP-1 (red) or DAPI nuclear stain (blue). This analysis was performed on three independently isolated C2C12 CLP-1 +/− cell cultures. (E) Graphical analysis of immunofluorescence of C2C12 CLP-1 +/+ (WT) and +/− cells in growth media (GM) and differentiation media (DM) for 0, 24, 48 and 72 hours. Antibodies to PCNA and myosin heavy chain were used, and DAPI was used as a nuclear stain. At each time point, cells were counted (n=500) and the ratio of cells expressing PCNA or myosin heavy chain versus total cells is depicted graphically (n=3).