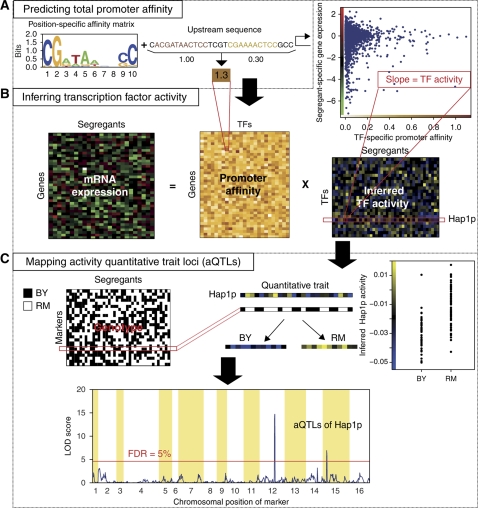
Figure 1.
Overview of our transcription-factor-centric approach to detecting trans-acting sequence variation. (A) We construct a matrix containing the promoter-binding affinity for each combination of the upstream non-coding sequence of a particular gene and the position-specific affinity matrix (PSAM) of a particular transcription factor (TF). (B) The promoter-binding affinity matrix is interpreted as a regulatory connectivity matrix and used to infer a matrix containing the regulatory activity of each TF in each segregant. For each segregant independently, multivariate genome-wide linear regression of segregant-specific differential mRNA expression on the matrix of promoter affinity for all TFs is performed. The coefficients from this linear fit represent (differential) protein-level TF activities. (C) For each TF independently, we treat the inferred activity as a quantitative phenotype and use genetic linkage analysis across all segregants to identify loci that genetically modulate TF activity. Whenever TF activity is statistically associated with genotype at a particular genetic marker, this shows as a high log-odds (LOD) score indicating the presence of a TF activity quantitative trait locus, or ‘aQTL’.