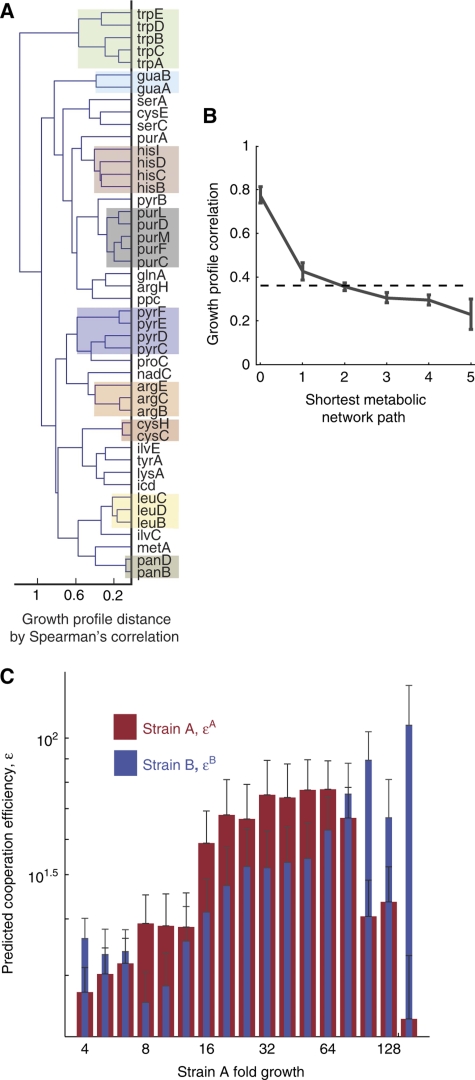
Figure 3.
SMIT interactions are emergent properties of the metabolic network. (A) A dendrogram depicts hierarchical clusters derived from measured cooperation profiles. Most of the major metabolic pathways are recovered as distinct branches, implying that functional relationships can be deduced from SMIT interactions. Co-phenetic correlation: 0.71. (B) Mean growth profile correlations are plotted against metabolic network distance. The correlation between growth profiles declines with the distance spanning mutations. The dotted line shows the mean correlation for all pairs. Error bars are 95% bootstrap confidence intervals. (C) Theoretical cooperation efficiencies predict the features of cooperative growth. Mean cooperation efficiencies are presented for sets of co-cultures binned by the measured growth of strain A. Cooperation efficiencies, ɛ, are derived a priori from stoichiometric models with no free parameters. A high cooperation efficiency predicts that a strain can cheaply produce metabolites that effect high growth in the conjugate partner. A's growth tends to increase monotonically with ɛB, B's cooperation efficiency, but reaches a maximum for intermediate levels of ɛA, consistent with the invested benefits model of cooperation.