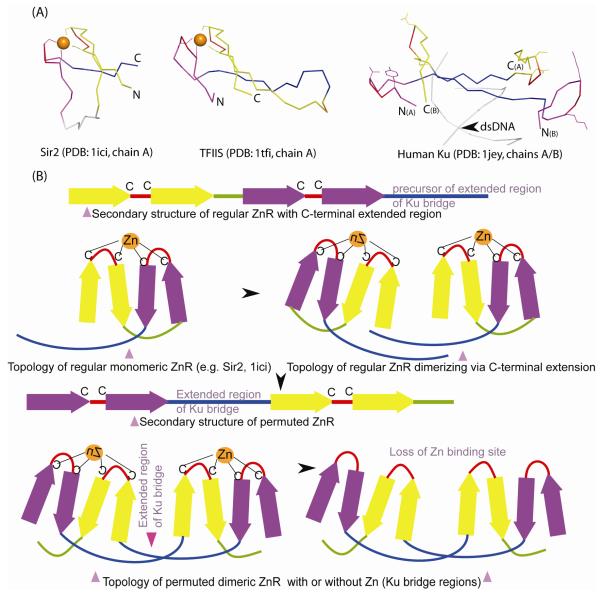
Figure 3.
Structural comparison of Ku bridge-region to Zn-ribbons.
(A) Structural diagrams of Zn-ribbons from Sir2 (left; 1ici: chain B, 119-156), TFSII (middle; 1tfi: chain A, 8-50) and bridge-region of the human Ku heterodimer (right; 1jey: chain A, 287-313; chain B, 277-307) are shown. The structures were superimposed using the program PyMOL by manually defining equivalent residue pairs from regions comprising the zinc knuckles (red, purple) and the β-strands (yellow, blue) and translated for clarity. In some of the alignments, circular permutation needs to be assumed in order to obtain best structural superimposition. Structurally equivalent regions are colored similarly. The zinc chelating residues of Sir2 and TFSII and the equivalent residues from Ku are shown as ball-and-sticks. The zinc ion is indicated as an orange sphere. The primary β-hairpin of the Zn-ribbon is colored purple, the extended region of Ku-bridge in blue, the zinc knuckle in red and the other β-strands in yellow.
(B) A permutation scheme for arriving at the Ku bridge-region structure from a Sir2-like precursor. In its ancestral state, Ku Zn-ribbons, resembling Sir2, likely dimerized via their C-terminal extended regions. A circular permutation resulted in placing the extended region of Ku-bridge between the primary and secondary zinc knuckles (colored purple and yellow) and extensive hydrogen-bonding interactions likely locked this atypical conformation in place. This is then likely to have been followed by the loss of Zn-binding sites, to abolish zinc dependence of NHEJ.