Figure 1.
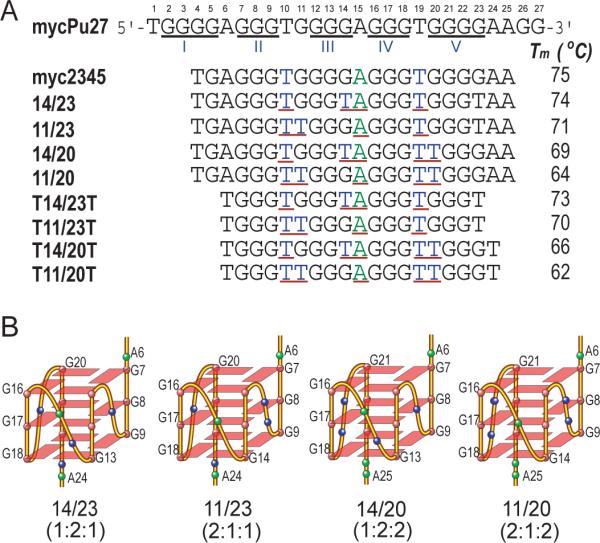
(A) The promoter sequence of the NHE III1 element of the c-Myc gene and its modifications. MycPu27 is the wild-type 27-mer G-rich sequence of the c-Myc NHE III1. The five runs of guanines are underlined and numbered from 5'-end. The numbering system is shown above mycPu27. Myc2345 is the 22-nt G-rich sequence that adopts the major c-Myc promoter G-quadruplex. Myc2345 has four loop-isomers, which can be isolated by dual G-to-T substitutions at (14,23), (11,23), (14,20), and (11,20) positions, namely, 14/23, 11/23, 14/20, and 11/20, respectively. 14/23 is the mutant Myc2345 that forms the major loop-isomer and was used for NMR structure determination (26). The truncated sequences of four loop-isomers have only one flanking T at each end. The three loops of each loop-isomer are underlined. The melting temperatures (Tm) of all the sequences in 20 mM K+ solution are also shown. (B) Schematic drawing of the folding topologies of the four loop-isomers of the major c-Myc G-quadruplex Myc2345 in K+ solution. The four loop-isomers all adopt parallel-stranded folding and have different arrangements of 1- and 2-nt loops. Base colors: red = guanine, blue = thymine, green = adenine.
