Abstract
With eukaryotic non-coding RNAs (ncRNAs) now established as critical regulators of cellular transcription, the true diversity with which they can elicit biological effects is beginning to be appreciated. Two ncRNAs, mouse B2 RNA and human Alu RNA, have been found to repress mRNA transcription in response to heat shock. They do so by binding directly to RNA polymerase II, assembling into complexes on promoter DNA, and disrupting contacts between the polymerase and the DNA. Such a mechanism of repression had not previously been observed for a eukaryotic ncRNA; however, there are examples of eukaryotic protein domains that repress transcription by blocking essential protein-DNA interactions. Comparing the mechanism of transcriptional repression utilized by these protein domains to that used by B2 and Alu RNAs raises intriguing questions regarding transcriptional control, and how B2 and Alu RNAs might themselves be regulated.
Keywords: ncRNA, RNA polymerase II, transcription, SINE
Within the past several years a rising number of non-coding RNAs (ncRNAs) have been found to function as regulators of RNA polymerase II (Pol II) transcription. Although ncRNAs have long been recognized as central participants in controlling other stages of gene expression, such as mRNA splicing and translation, their emergence as direct regulators of mRNA transcription is still relatively novel. ncRNA regulators of mRNA transcription can be loosely categorized into two groups: those that facilitate chromatin modifications and those that control the activity of a transcription factor.1 Of the ncRNAs in the latter category (e.g. those that bind to transcriptional activators, coactivators, or the general transcription machinery), it is of particular interest that they do so via diverse mechanisms, and function in diverse biological systems. This suggests that we only understand a small subset of the ncRNA transcriptional regulators that likely exist in eukaryotic cells. The realization that much of the genome is transcribed2,3 only bolsters this postulation.
For a subset of eukaryotic ncRNA transcriptional regulators, biochemical experiments have revealed detailed mechanisms by which they function. This is true of two ncRNAs, mouse B2 RNA and human Alu RNA, which we have found to use a common mechanism to repress transcription, involving direct binding of the ncRNAs to Pol II.4-6 Here we will compare the mechanism of transcriptional repression employed by B2 and Alu RNAs to mechanisms used by protein domains in two multi-subunit eukaryotic general transcriptional factors, SNAPc and TFIID. Drawing these comparisons raises several intriguing questions regarding transcriptional control and how B2 and Alu RNAs might themselves be regulated.
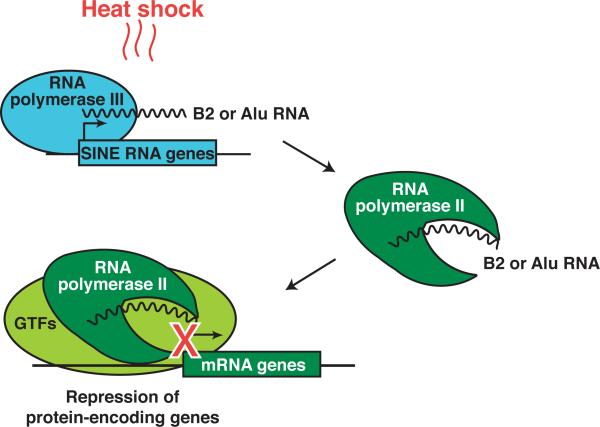
Mouse B2 RNA and human Alu RNA are transcribed as discrete transcripts by RNA polymerase III (Pol III) from short interspersed elements (SINEs) that are widely abundant within their respective genomes.7 B2 RNA is approximately 180 nt in length, and Alu RNA is approximately 280 nt in length. Upon cellular stress, such as heat shock, the abundance of B2 and Alu RNAs sharply increases in mouse and human cells, respectively.8,9 Our lab has shown that in response to heat shock, B2 RNA and Alu RNA bind to Pol II and repress transcription of several protein encoding genes (Figure 1).4-6 This repression is in contrast to the activation of heat shock stimulated genes, such as hsp70. These are the first examples of mammalian ncRNAs that bind to Pol II, and their roles as transcriptional repressors provide a biological function for SINEs, which have historically been considered junk DNA.
Figure 1.
Mouse B2 RNA and human Alu RNA are repressors of Pol II transcription. Schematic illustrating transcriptional repression by the SINE encoded ncRNAs B2 (mouse) and Alu (human). These ncRNAs are transcribed by Pol III, and their levels increase upon heat shock. B2 and Alu RNA bind to Pol II and repress transcription of select mRNA genes. These ncRNAs incorporate into complexes at promoters along with Pol II and general transcription factors (GTFs). Biochemical experiments show that the ncRNAs disrupt contacts between Pol II and the DNA, and are most consistent with a model in which the repression domains of B2 and Alu RNAs bind in the DNA cleft of Pol II. See main text for references.
We have studied the mechanism by which B2 RNA and Alu RNA function as transcriptional repressors, taking advantage of a highly purified, reconstituted Pol II transcription system. These experiments revealed that B2 RNA and Alu RNA bind directly to core Pol II with high affinity (KD < 2 nM), and that these complexes are kinetically stable (t1/2 > 1 hr).5,6 Interestingly, the association of B2 RNA or Alu RNA with Pol II does not block the assembly of the polymerase into complexes at promoter DNA. Rather, B2 RNA and Alu RNA can incorporate along with Pol II and general transcription factors into complexes on DNA.5,6 These complexes are transcriptionally inert due to the presence of the ncRNA. Although this mechanism of repression was initially revealed by in vitro biochemical experiments, it has since been corroborated by cell based studies. In heat shocked mouse or human cells, B2 RNA or Alu RNA are found with Pol II at the promoters of transcriptionally repressed genes.6
We have explored in greater detail precisely how B2 RNA and Alu RNA prevent transcription from occurring once they assemble into complexes at promoters by studying the network of protein-DNA interactions that occur within these complexes. We found that in inhibited complexes on promoter DNA, B2 RNA and Alu RNA disrupted contacts between Pol II and the DNA throughout the core promoter.10 First, the ncRNAs eliminated the crosslinking typically observed between promoter DNA and Pol II in preinitiation complexes. Interestingly, however, crosslinking between promoter DNA and the general transcription factor IIB (TFIIB) was enhanced by the presence of B2 RNA and Alu RNA, suggesting that the presence of a repressor ncRNA alters the conformation of complexes. Second, in DNase I footprinting experiments, the presence of B2 RNA or Alu RNA eliminated nearly all the protection that is ascribed to Pol II in preinitiation complexes, while protections and enhancements due to the TATA binding protein (TBP) and TFIIB were maintained.10 Together these experiments support a model for transcriptional repression in which B2 RNA and Alu RNA enter assembling complexes at promoters and prevent Pol II from properly engaging the DNA, thereby altering the conformation of complexes and blocking RNA synthesis (Figure 1). Pol II is likely held on the DNA within these inactive complexes through its interactions with promoter-bound TBP and TFIIB.
This mechanism for repressing eukaryotic transcription by ncRNAs is unique; however, analogies can be drawn to proteins that block DNA binding events in order to regulate eukaryotic transcription. For example, the transcription factor SNAPc (snRNA activating protein complex) is regulated by a protein domain that blocks DNA binding (Figure 2A).11 SNAPc is a transcription factor composed of five protein subunits that binds to an element in Pol II and Pol III snRNA promoters.12 A partial SNAPc complex that lacked both the C-terminal region of the SNAP190 subunit and the SNAP45 subunit bound DNA much more efficiently than the complete SNAPc complex.11 This suggested that the C-terminal region of SNAP190 and/or SNAP45 serve as an internal damper to repress DNA binding by SNAPc. Interestingly, the C-terminal region of SNAPc binds directly to the transcriptional activator Oct-1.13 Oct-1 binding to the C-terminal region of SNAP190 allows the SNAPc complex to properly bind DNA, thus Oct-1 relieves the repression caused by the internal damper within SNAPc.11
Figure 2.
Subunits of the SNAPc and TFIID complexes contain domains that function as internal dampers of DNA binding. (A) A model for how DNA binding by SNAPc is regulated. The C-terminal region of SNAP190 and/or SNAP45 represses DNA binding by the SNAPc complex. De-repression occurs when the transcriptional activator Oct-1 binds to the C-terminal domain of SNAP190, perhaps triggering a conformational change that converts the dampers of DNA binding into handles that stabilize the complex with Oct-1. (B) Schematic illustrating a model for how DNA binding by TFIID is de-repressed by the transcriptional activator cJun. The N-terminal region of TAF1 interacts with the concave surface of TBP, which represses TFIID binding to DNA. Direct interactions between cJun and the N-terminal region of TAF1 destabilize the TAF1-TBP contacts, allowing TBP to bind to the TATA box. See main text for references.
Similar to the SNAPc example is the general transcription factor TFIID, which also contains a subunit that dampens the ability of TFIID to bind DNA. TFIID is a large complex consisting of TBP, which binds to TATA boxes, and 13 TAFs (TBP associated factors), which can associate with other core promoter elements.14 TFIID nucleates Pol II preinitiation complex formation at the promoters of genes. The N-terminus of TAF1 has been shown to interact with the DNA-binding surface of TBP, thereby repressing DNA binding by TFIID (Figure 2B).15,16 Other factors have been shown to interact with TFIID to de-repress the TAF1-mediated inhibition of TBP-DNA binding (albeit by different mechanisms); these factors include transcription factor IIA (TFIIA)17,18, the transcriptional activator cJun,19,20 and the acidic activation domain of viral protein 16 (VP16).21
Parallels can be drawn between the internal dampers of DNA binding in SNAPc and TFIID, and the ncRNA repressors B2 RNA and Alu RNA. B2 and Alu RNAs act similarly to the C-terminal region of SNAP190 and/or SNAP45 and the N-terminal region of TAF1 – they all prevent a multi-protein complex from binding DNA, thereby repressing transcription. In the case of TAF1, structural data has provided clarity on the mechanism by which this occurs. A solution structure showed that the N-terminal region of TAF1 folds in a manner that loosely mimics the surface of TATA DNA, thereby allowing TAF1 to occupy the concave DNA-binding surface of TBP.22 Exactly how the internal damper in SNAPc elicits its effects is not known11; two possible mechanisms include: competing with DNA binding by interacting with the surface of SNAPc that binds DNA, or causing a conformational change within SNAPc that occludes DNA binding. As for B2 and Alu RNA, both of these mechanisms are also possible; however, we favor the former mechanism (i.e. the ncRNAs bind Pol II in a manner that blocks DNA binding). Although we do not know precisely where on Pol II these ncRNAs bind, insight can be obtained from a crystal structure of yeast Pol II bound to a synthetic RNA aptamer (named Fc) that represses transcription.23 The Fc aptamer occupies the DNA binding cleft of Pol II in a manner that would preclude the template DNA strand from entering the cleft. Our data are consistent with the regions of B2 RNA and Alu RNA that mediate transcriptional repression (named repression domains) interacting with the DNA cleft of Pol II in a mode similar to the synthetic Fc aptamer.10,24 Presuming this is the case, then B2 RNA and Alu RNA repression domains interact with Pol II much like the TAF1 subunit of TFIID interacts with TBP (i.e. to the DNA binding surface), thereby competing with DNA binding. Perhaps this manner of binding – masking a functional region on TFIID or Pol II as opposed to binding to an outside surface and inducing an overall conformational change – allows TAF1 and B2/Alu RNA to meet with the most success as negative regulators.
Drawing parallels between the aforementioned protein domains and B2 and Alu RNAs provides a framework for contemplating whether their similar mechanisms of regulation extend beyond controlling transcription of genes. In the case of SNAPc, it was proposed that the internal damper might prevent it from associating with non-specific sites in the genome – especially important because this factor does not exhibit strong sequence specificity for binding DNA, but dissociates from DNA slowly.11 Analogous properties have been observed with TBP alone (i.e. stable association with non-specific DNA).25 Hence, in the TFIID complex, the N-terminus of TAF1 could refine the DNA accessibility of its TBP subunit.15-18 Therefore, in both cases the inhibitory protein domains may prevent the transcription factors from binding indiscriminately to DNA. Perhaps non-specific DNA binding by Pol II can also be controlled by molecules that bind in its DNA cleft; ncRNAs could play this role and prevent non-productive interactions between the polymerase and the genome from occurring. Because we know that B2 RNA and Alu RNA can prevent Pol II from engaging the promoter at repressed genes during heat shock, these ncRNAs could serve in general to limit the access of Pol II to DNA under conditions of cellular stress. Moreover, it is conceivable that unidentified ncRNAs bind the cleft to generally prevent non-specific polymerase-genome interactions.
When drawing comparisons between eukaryotic ncRNA transcriptional repressors and protein domains that repress transcription via similar mechanisms, one critical gap becomes apparent. In the cases of SNAPc and TFIID, mechanisms clearly exist to counteract the effects of their inhibitory domains. Specific transcriptional activator proteins have been found to bind directly to the inhibitory regions and de-repress DNA binding, thereby mediating transcriptional activation – Oct-1 binds to the C-terminal region of SNAP19011, and cJun bind to the N-terminal region of TAF1.19,20 In these models for de-repression, the activator proteins are thought to simply move the inhibitory domain out of the way to allow DNA binding. Perhaps cellular factors exist that bind to B2 RNA and Alu RNA repression domains to move them out of the DNA binding cleft of Pol II in order to de-repress transcription. Alternatively, a factor might completely dissociate B2 RNA and Alu RNA from Pol II – a factor could do so via contacting the ncRNAs themselves or the polymerase. Because in the case of Pol II the inhibitor is an RNA, it is also conceivable that an RNase could clip or degrade the ncRNAs either within inhibited complexes or from Pol II prior to its association with DNA, thereby de-repressing the polymerase. Biochemical evidence has shown that degrading B2 RNA or Alu RNA by RNase treatment in promoter-bound complexes re-activates the complexes such that transcription can occur.5,6 Therefore, inhibition by these ncRNAs is reversible, which is an important feature of their repression. It is clear that one or more mechanisms must be present in cells to regulate transcriptional repression by B2 and Alu RNAs such that genes such as hsp70 can be activated upon heat shock and transcription of repressed genes can resume as cells recover from heat shock. The mechanisms of de-repression for the ncRNAs, and the cellular factors involved, still await discovery.
We have focused on eukaryotic factors in drawing parallels between ncRNAs and protein domains that disrupt protein-DNA interactions; however, it is also worth mentioning prokaryotic 6S RNA because it too disrupts a protein-DNA interaction – that between E. coli RNA polymerase and promoter DNA. Bacterial RNA polymerases (RNAPs) contain a dissociable σ subunit that interacts with promoter DNA elements to facilitate promoter recognition and melting of the DNA to form open complexes.26 In E. coli, σ70 predominates during exponential growth.27 As cells enter stationary phase, 6S RNA accumulates and binds σ70-RNAP to repress transcription at σ70 promoters.28 Transcriptional de-repression can occur by 6S RNA serving as a template for RNA-dependent RNA synthesis of short de novo transcripts.29,30 Upon synthesis of these short transcripts, the σ70 subunit releases and the remaining complex between core RNAP and 6S RNA is unstable.
6S RNA is thought to repress transcription by mimicking promoter DNA, thus blocking the polymerase from binding. The 6S RNA secondary structure contains a bulge that is analogous to the conformation of melted promoter DNA,31,32 and indeed, in vitro studies revealed that 6S RNA blocks σ70-RNAP from binding to a promoter.30 More recent studies suggest, however, that the interplay of 6S RNA, promoter DNA, and σ70-RNAP might be more complex. First, inhibition by 6S RNA is promoter specific – all σ70 promoters are not repressed.33 Second, the regions of σ70 that interact with 6S RNA are overlapping but distinct from those that interact with promoter DNA, which might contribute to the promoter specificity observed for transcriptional repression.34
When comparing the mechanism of transcriptional repression by 6S RNA to that of the B2 and Alu RNAs, similarities and differences emerge. The most apparent similarities are that all these ncRNAs bind directly to RNA polymerases and interfere with promoter binding. In the case of 6S RNA, it does not appear that complexes containing the ncRNA, the promoter, and the polymerase form. By contrast, in the case of B2 RNA and Alu RNA, such complexes form both in vitro and in cells. In the eukaryotic system, it is likely that interactions between Pol II and general transcription factors allow promoter-bound complexes containing inhibitory ncRNA to assemble despite the disrupted contacts between Pol II and the DNA. As discussed earlier, how transcriptional repression by B2 RNA and Alu RNA is alleviated awaits discovery. It is possible that, like 6S RNA, de-repression might involve RNA dependent RNA polymerization; extension of the 3' end of RNA by yeast Pol II has been observed.35
Much remains to be learned about precisely how B2 RNA and Alu RNA dock to Pol II and repress transcription, and importantly, how this process is regulated. Drawing comparisons between these ncRNAs and eukaryotic inhibitory protein domains, as well as prokaryotic 6S RNA, provides insight into B2 and Alu RNAs’ functions. With further study of both ncRNA and protein transcriptional regulators it will become apparent whether repressing the DNA binding activity of eukaryotic general transcription factors and polymerases is a more universal mechanism of regulation.
Acknowledgements
We apologize to those colleagues whose work was not described due to space limitations. Our work on ncRNA repressors of Pol II has been supported by Public Health Service grant (R01 GM068414) from the National Institute of General Medical Sciences.
Abbreviations used
- ncRNA
non-coding RNA
- Pol II
RNA polymerase II
- Pol III
RNA polymerase III
- SNAPc
snRNA activating protein complex
- TAF
TATA binding protein associated factor
- TBP
TATA binding protein
- TFII-
transcription factor of RNA polymerase II
- VP16
viral protein 16
References
- 1.Goodrich JA, Kugel JF. From bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Crit Rev Biochem Mol Biol. 2009;44:3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–23. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 3.Pheasant M, Mattick JS. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–53. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- 4.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–21. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 5.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–9. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 6.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 7.Kramerov DA, Vassetzky NS. Short retroposons in eukaryotic genomes. Int Rev Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- 8.Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–72. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 9.Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucl Acids Res. 1995;23:1758–65. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci USA. 2009;106:5569–74. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittal V, Ma B, Hernandez N. SNAP(c): a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev. 1999;13:1807–21. doi: 10.1101/gad.13.14.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J Biol Chem. 2001;276:26733–6. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 13.Mittal V, Cleary MA, Herr W, Hernandez N. The Oct-1 POU-specific domain can stimulate small nuclear RNA gene transcription by stabilizing the basal transcription complex SNAPc. Mol Cell Biol. 1996;16:1955–65. doi: 10.1128/mcb.16.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–79. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 15.Kokubo T, Yamashita S, Horikoshi M, Roeder RG, Nakatani Y. Interaction Between the N-Terminal Domain of the 230-kDa Subunit and the TATA Box-Binding Subunit of TFIID Negatively Regulates TATA-Box Binding. Proc Natl Acad Sci USA. 1994;91:3520–4. doi: 10.1073/pnas.91.9.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokubo T, Gong D-W, Yamashita S, Horikoshi M, Roeder RG, Nakatani Y. Drosophila 230-kD TFIID subunit, a functional homolog of the human cell cycle gene product, negatively regulates DNA binding of the TATA box-binding subunit of TFIID. Genes Dev. 1993;7:1033–46. doi: 10.1101/gad.7.6.1033. [DOI] [PubMed] [Google Scholar]
- 17.Kokubo T, Swanson MJ, Nishikawa JI, Hinnebusch AG, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–12. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozer J, Mitsouras K, Zerby D, Carey M, Lieberman PM. Transcription factor IIA derepresses TATA-binding protein (TBP)- associated factor inhibition of TBP-DNA binding. J Biol Chem. 1998;273:14293–300. doi: 10.1074/jbc.273.23.14293. [DOI] [PubMed] [Google Scholar]
- 19.Lively TN, Ferguson HA, Galasinski SK, Seto AG, Goodrich JA. c-Jun binds the N terminus of human TAF(II)250 to derepress RNA polymerase II transcription in vitro. J Biol Chem. 2001;276:25582–8. doi: 10.1074/jbc.M100278200. [DOI] [PubMed] [Google Scholar]
- 20.Lively TN, Nguyen TN, Galasinski SK, Goodrich JA. The basic leucine zipper domain of c-Jun functions in transcriptional activation through interaction with the N terminus of human TATA-binding protein-associated factor-1 (human TAF(II)250). J Biol Chem. 2004;279:26257–65. doi: 10.1074/jbc.M400892200. [DOI] [PubMed] [Google Scholar]
- 21.Kotani T, Banno K, Ikura M, Hinnebusch AG, Nakatani Y, Kawaichi M, Kokubo T. A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding of transcription factor IID. Proc Natl Acad Sci USA. 2000;97:7178–83. doi: 10.1073/pnas.120074297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D, Ishima R, Tong KI, Bagby S, Kokubo T, Muhandiram DR, Kay LE, Nakatani Y, Ikura M. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell. 1998;94:573–83. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 23.Kettenberger H, Eisenfuhr A, Brueckner F, Theis M, Famulok M, Cramer P. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat Struct Mol Biol. 2006;13:44–8. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- 24.Espinoza CA, Goodrich JA, Kugel JF. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA. 2007;13:583–96. doi: 10.1261/rna.310307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman RA, Pugh BF. Evidence for the functional binding and stable sliding of the TATA binding protein on nonspecific DNA. J Biol Chem. 1995;270:13850–9. doi: 10.1074/jbc.270.23.13850. [DOI] [PubMed] [Google Scholar]
- 26.Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Mol Cell. 2005;20:335–45. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–66. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 28.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–23. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 29.Gildehaus N, Neusser T, Wurm R, Wagner R. Studies on the function of the riboregulator 6S RNA from E. coli: RNA polymerase binding, inhibition of in vitro transcription and synthesis of RNA-directed de novo transcripts. Nucleic Acids Res. 2007;35:1885–96. doi: 10.1093/nar/gkm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wassarman KM, Saecker RM. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science. 2006;314:1601–3. doi: 10.1126/science.1134830. [DOI] [PubMed] [Google Scholar]
- 31.Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–84. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol. 2005;12:313–9. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- 33.Cavanagh AT, Klocko AD, Liu X, Wassarman KM. Promoter specificity for 6S RNA regulation of transcription is determined by core promoter sequences and competition for region 4.2 of sigma70. Mol Microbiol. 2008;67:1242–56. doi: 10.1111/j.1365-2958.2008.06117.x. [DOI] [PubMed] [Google Scholar]
- 34.Klocko AD, Wassarman KM. 6S RNA binding to Esigma(70) requires a positively charged surface of sigma(70) region 4.2. Mol Microbiol. 2009;73:152–64. doi: 10.1111/j.1365-2958.2009.06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehmann E, Brueckner F, Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450:445–9. doi: 10.1038/nature06290. [DOI] [PubMed] [Google Scholar]