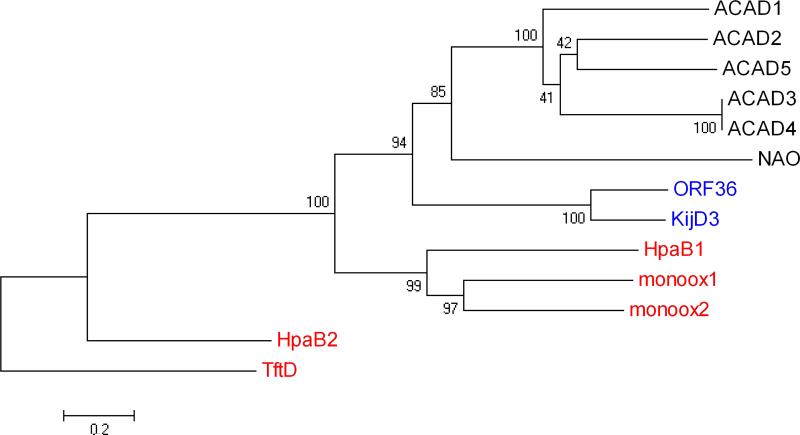
Figure 6.
Evolutionary relationships of 13 taxa including nitrososynthases, acyl-CoA dehydrogenases, and flavin-containing monooxygenases. The evolutionary history was inferred using the Neighbor-Joining method (42). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (43). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (43). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (44) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 333 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (45). The nitrososynthases ORF36 and KijD3 are highlighted in blue text, the Class D flavin-containing monooxygenases in red, and the acyl-CoA dehydrogenases and nitroalkane oxidase are in black text. Proteins included here (which are also included in Supporting Information, Table S1) are named as follows: ACAD1, short branched-chain acyl-CoA dehydrogenase (H. sapiens), 2JIF (35); ACAD2, butyryl-CoA dehydrogenase (M. elsedenii), PDB entry 1BUC (58); ACAD3 medium-chain acyl-CoA dehydrogenase (S. scrofa), PDB entry 3MDE (59); ACAD4, medium-chain acyl-CoA dehydrogenase (S. scrofa), PDB entry 1UDY (60); ACAD5, short-chain acyl-CoA dehydrogenase (R. norvegicus), PDB entry 1JQI (61); HpaB1, 4-hydroxyphenylacetate monooxygenase (A. baumannii), PDB entry 2JBT (24); monoox1, 3-hydroxy-9,10-seco-nandrost-1,3,5(10)-triene-9,17-dione hydroxylase (Rhodococcus sp Rha1), PDB entry 2RFQ (62); monoox2, putative hydroxylase (Rhodococcus sp Rha1), PDB entry 2OR0 (63); NAO, nitroalkane oxidase (F. oxysporum), PDB entry 2C0U (64); HpaB2, 4-hydroxyphenylacetate monooxygenase (T. thermophilus), PDB entry 2YYI (25); TftD, chlorophenol-4-monooxygenase (B. cepacia), PDB entry 3HWC (26).