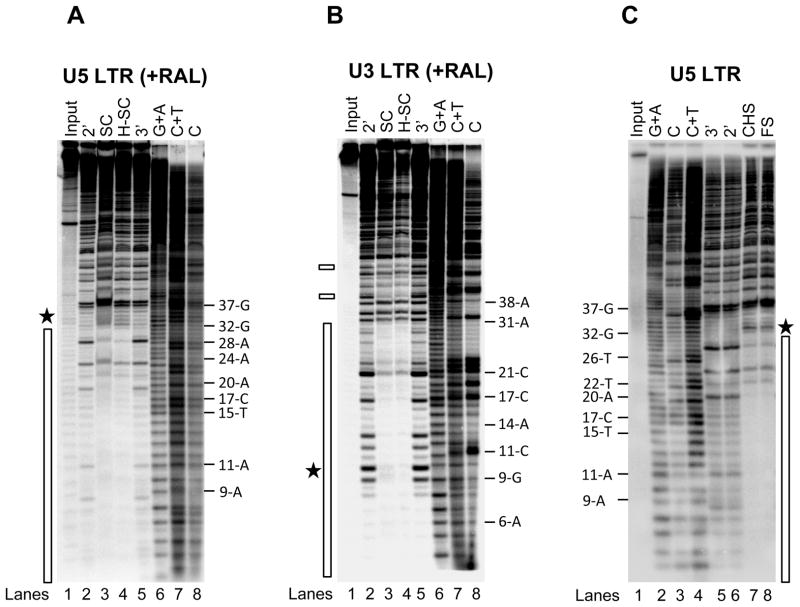
Figure 4.
RAL does not affect the IN multimeric structure at the LTR ends. (A) IN (60 nM) was pre-assembled with 5′-32P end-labeled 1.6 kb U5 blunt-ended DNA (3 nM) for 15 min at 14°C. RAL (750 nM) and supercoiled DNA (3 nM) were added and incubated for 2 h at 37°C. Samples were treated with DNaseI and subjected to native agarose gel electrophoresis. DNA was purified from the trapped SC and H-SC and subjected to denaturing polyacrylamide 15% (w/v) gel electrophoresis. Lane 1, input DNA; lanes 2 and 5, input DNA digested with DNaseI for 2 min and 3 min, respectively; lanes 3 and 4, DNaseI treated SC and H-SC, respectively; lanes 6 to 8, Maxam-Gilbert chemical sequence markers prepared from input DNA. The nucleotide positions are marked on the right. The asterisk indicates enhanced DNaseI digestion and the vertical bar represents the protected DNA region. (B) DNaseI footprint analysis on 2.4 kb U3 blunt-ended DNA. The formation of trapped complexes, DNaseI treatment and sample loading order were identical as described above in A. Nucleotide positions on U3 LTR are on the right. Chemical sequence markers of U3 DNA are in lanes 6 to 8. The vertical bar represents the DNaseI protected region and two minor protected regions beyond 38 bp are marked by small rectangles on the left. (C) IN (80 nM) was pre-assembled with 5′-32P end-labeled 1.6 kb U5 blunt-ended DNA (3 nM) for 15 min at 14°C. Strand transfer was initiated by adding supercoiled DNA (3 nM) and incubating at 37°C for 2 h. Samples were treated with DNaseI, deproteinized and subjected to agarose gel electrophoresis. DNA purified from FS and CHS products were subjected to denaturing polyacrylamide 15% (w/v) gel electrophoresis. Lane 1, input DNA; lanes 2 to 4 Maxam-Gilbert chemical sequence markers prepared from input DNA; lane 5 and 6 contain input DNA digested with DNaseI for 3 min and 2 min, respectively; lanes 7 and 8, DNaseI treated CHS and FS, respectively. The nucleotide positions are marked on the left. The asterisk indicates enhanced DNaseI digestion and the vertical bar represents the protected DNA region.