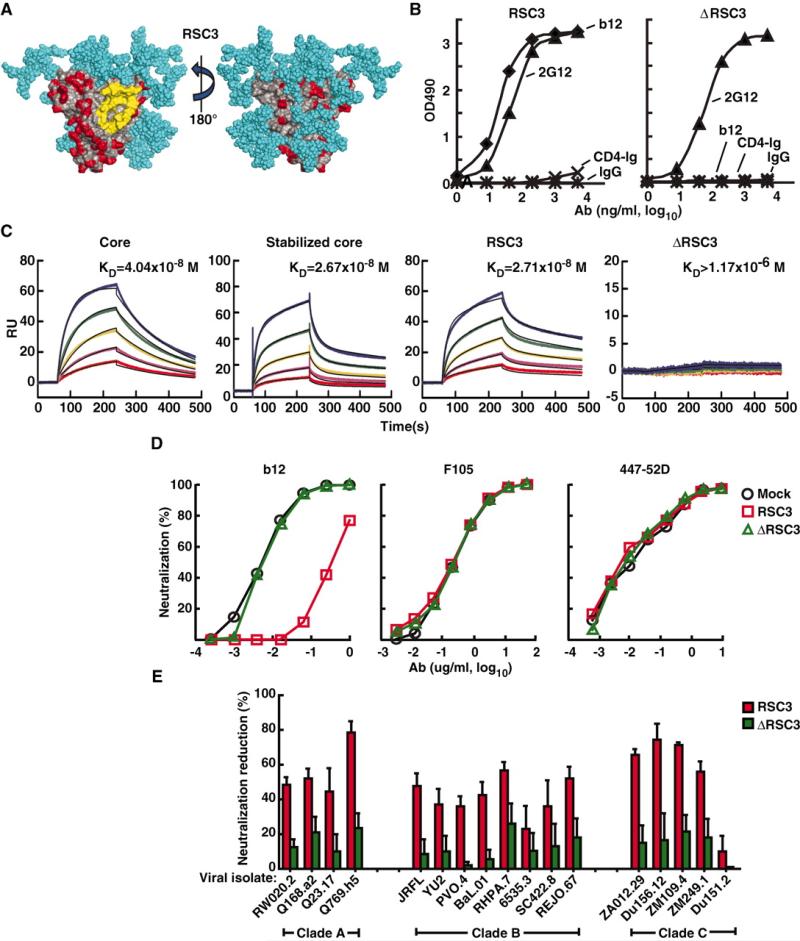
Fig. 1.
Design and antigenic profile of RSC3 and analysis of epitope-specific neutralization. (A) Surface structure model of the RSC3. The outer domain contact site for CD4 is highlighted in yellow. Regions highlighted in red are antigenically resurfaced areas, shown on both the inner (left) and outer (right) faces of the core protein. Glycans are shown in light blue. (B) Antigenicity of the RSC3 protein based on ELISA using the neutralizing CD4bs mAb b12 and CD4-Ig fusion protein. mAb 2G12 was used to confirm the structural integrity of the protein. OD indicates optical density. IgG, irrelevant IgG. (C) mAb b12 was immobilized on the sensor chip for SPR kinetic binding analysis with the proteins shown. RU, resonance units. (D) RSC3 blockade of HIV-1 viral strain HXB2 neutralization by the broadly neutralizing CD4bs mAb b12 but not mAb F105, which recognizes the CD4bs differently and has limited neutralization breadth. The V3-neutralizing mAb 447-52D is shown as a control. (E) Analysis of serum 45 neutralization of a panel of 17 viruses, using RSC3 and ΔRSC3 to block neutralization activity. The percent reduction in the serum 50% inhibitory dilution (ID50) caused by competition with RSC3 or ΔRSC3 is shown on the y axis (±SEM of three independent experiments). Viral strains and clades are shown on the x axis. Values less than 20% are not considered significant in this assay.