Figure 1.
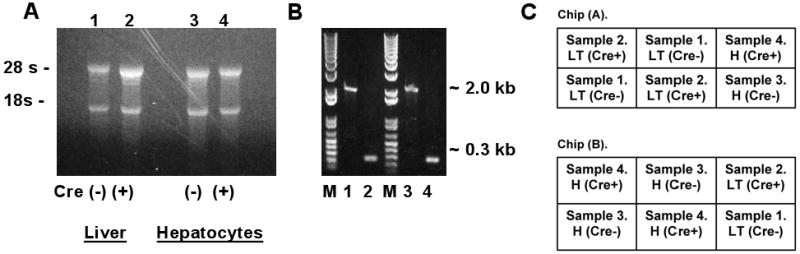
Analysis of RNA and DNA from male Cre(-) IkkβF/F and Cre(+) IkkβΔhep mouse livers and freshly isolated hepatocytes used for microarray profiling. (A) EtBr-stained RNA-sizing gel. (B) Deletion analysis. EtBr-stained DNA-sizing gel. Genotyping for deletion confirmation was performed by PCR using standard primers (Koch et al., 2009). M, size markers (bps); lanes #1-#4 as in panel (A); Cre(-) ∼2-kb, Cre(+) ∼0.3-kb. (C) Hybridization format: chips 1 (A) and 2 (B). Each RNA sample (liver tissue [LT], or isolated hepatocytes [H]) was obtained from a separate mouse: Samples #1 and #2, Cre(-)- and Cre(+)-tissues; #3 and #4, Cre(-)- and Cre(+)-isolated hepatocytes. Each sample was hybridized 3×, on 3 separate sectors across the 2 chips, as diagrammed.
