Abstract
Accommodation of donor and acceptor substrates is critical to the catalysis of (thio)phosphoryl group transfer, but there has been no systematic study of donor nucleotide recognition by kinase ribozymes, and there is relatively little known about the structural requirements for phosphorylating internal 2′OH. To address these questions, new self-phosphorylating ribozymes were selected that utilize ATP(gammaS) or GTP(gammaS) for 2′OH (thio)phosphorylation. Eight independent sequence families were identified among 57 sequenced isolates. Kinetics, donor nucleotide recognition and secondary structures were analyzed for representatives from each family. Each ribozyme was highly specific for its cognate donor. Competition assays with nucleotide analogs showed a remarkable convergence of donor recognition requirements, with critical contributions to recognition provided by the Watson–Crick face of the nucleobase, lesser contributions from donor nucleotide ribose hydroxyls, and little or no contribution from the Hoogsteen face. Importantly, most ribozymes showed evidence of significant interaction with one or more donor phosphates, suggesting that—unlike most aptamers—these ribozymes use phosphate interactions to orient the gamma phosphate within the active site for in-line displacement. All but one of the mapped (thio)phosphorylation sites are on unpaired guanosines within internal bulges. Comparative structural analysis identified three loosely-defined consensus structural motifs for kinase ribozyme active sites.
INTRODUCTION
The RNA world hypothesis postulates a primitive RNA-directed metabolism before the evolution of genetically encoded protein synthesis [for a review see ref. (1)]. This notion has received support from recognition that the peptidyl transferase center of the bacterial ribosome is composed completely of RNA (2), from the discovery of eight natural classes of ribozymes that perform hydrolysis and phosphate ester exchange reactions (3), and from the identification of numerous riboswitches and non-coding RNAs (4,5). The RNA world hypothesis is also supported experimentally by the identification of artificial nucleic acid catalysts through in vitro selection methods. These catalysts promote a variety of chemical transformations, including amide bond formation (6), carbon–carbon bond formation (7), alkyl group transfer (8), acyl transfer (9), phosphodiester bond formation (10), limited RNA polymerization (11) and aminoacylation (12). Mapping the functional versatility and catalytic proficiency of ribozymes helps to constrain RNA world theories, while also generating tools that could potentially be used to re-engineer cellular metabolisms.
Phosphoryl transfer is a ubiquitous and important reaction in modern biology. It drives unfavorable reactions by generating chemically labile or conformationally unstable intermediates, it decreases metabolite permeability across membranes, it regulates protein–protein interactions and enzyme activity, it amplifies intracellular signals and it enables DNA synthesis and repair. Phosphoryl transfer is almost certainly one of the most ancient enzymatic activities and may pre-date the invention of genetically-encoded protein synthesis (1). Kinase activity is well established among catalytic nucleic acids selected in vitro. Kinase ribozymes able to transfer a (thio)phosphoryl group from an ATP(γS) donor to their 5′- or 2′-OH were first isolated by Lorsch and Szostak (13), and subsequently by the Burke and Bartel labs (14,15). One ribozyme, denoted 2PT3.1min, underwent multiple cycles of thiophosphoryl hydrolysis and re-thiophosphorylation (16), analogous to molecular motors that use ATP hydrolysis to power a work cycle. Catalytic DNA strands with kinase activity have also been described (17–20).
The chemistry of phosphoryl transfer is expected to be optimal when the acceptor nucleophile is close to the gamma phosphate and directly aligned with the nucleotide diphosphate leaving group on the donor. Nucleophilicity of the acceptor group is also likely to play a role; however, the observation that catalysis by two different kinase ribozymes was independent of pH (14,21) suggests that the primary role of the RNA is to reduce substrate entropy by bringing the donor and acceptor into alignment. The evolutionary and engineering challenges for generating new kinase ribozymes are therefore intimately associated with the challenge of constructing binding pockets that suitably organize the donor and acceptor substrates.
Nucleotide triphosphates (or their γ-thio analogs) serve as donors for all kinase ribozymes identified to date. There have been no systematic studies of donor nucleotide recognition by kinase ribozymes, and there is almost no information about how these ribozymes bind and orient the nucleotide donors. In contrast, numerous aptamers have been selected to bind nucleotide triphosphates and other nucleotide cofactors (22–33), and the details of their molecular interactions have been revealed largely through NMR analysis and computational modeling (34–38). These studies have shown that nucleotides are held in aptamer binding pockets primarily through base stacking and through hydrogen bonding with the sugar and with the Hoogsteen and Watson–Crick faces of the nucleobase, with little or no contribution from the phosphates. However, mere binding cannot be expected to predispose an RNA for catalysis (39). Formation of an active site that productively orients the donor nucleotide for catalysis may require more extensive or specialized interactions than those required for recognition by aptamers.
As for the phosphoryl acceptor moiety, the ribose hydroxyls are expected to offer the greatest opportunity for being organized into a productive conformation. The ribose hydroxyls of the RNA backbone present relatively low diffusional and conformational entropy as compared to a fully diffusible acceptor substrate. Kinase ribozyme selections have yielded species that (thio)phosphorylate various positions within the RNA chain, especially on the 5′OH or on internal 2′OH groups. Phosphorylation of 5′OH has been studied in some detail, particularly for the Kin.46 ribozyme (13,21,40). Ribozymes that kinase internal positions have remained largely uncharacterized.
To gain a better understanding of the structure–function relations in this class of ribozymes, we performed an in vitro selection for new kinase ribozymes capable of transferring a thiophosphoryl group from ATPγS or GTPγS to an internal 2′OH. Representatives from eight independently derived ribozyme families were analyzed to establish kinetic parameters for catalysis of phosphoryl transfer, to identify the determinants of substrate recognition, to establish their respective secondary structures, and to map thiophosphorylation sites. Comparative analysis identified a remarkable functional convergence that included ribozyme interactions with one or more phosphates in the donor nucleotide for nearly all the ribozymes and a non-random predilection for phosphorylating at guanosines.
MATERIALS AND METHODS
Materials
Oligodeoxynucleotides were purchased from Integrated DNA technologies (IDT, Coralville, IA). RNA was transcribed in vitro using phage T7 RNA polymerase, which was overproduced in bacteria and purified in the lab. Other enzymes were purchased from New England Biolabs (Ipswitch, MA), Ambion (Austin, TX) and Amersham Biotech (Pittsburgh, PA). ATPγS and GTPγS were purchased from Sigma (St. Louis). Radiolabeled nucleotides for 5′-, internal and 3′-labeling ([γ32P]-ATP, [α32P]-CTP and [α32P]-dATP, respectively) were purchased from Perkin-Elmer (Waltham, MA). N-acryloyl-aminophenylmercuric chloride (APM) was prepared as described (41).
Selection scheme
The initial RNA library (∼1014 unique species) was generated as described previously (14) with the following nucleotide sequence: 5′GGACCCUAGGGAAAAGCGAAUCAUACACAAGA(N70)GGGCAUGGUAUUUAAUUCCAUA 3′. During the selection, the first 8 nt were ligated to the library post-transcriptionally (14) to introduce a HEG-tethered deoxycytidine as an extra potential phosphorylation target. Because the selected species were found to be independent of this tethered substrate, full-length RNA was transcribed directly for all post-selection analysis.
During each selection cycle, gel-purified RNA was unfolded at 75–80°C for 5 min in water, followed by addition of 5× kinase reaction buffer (1× = 6 mM MgCl2, 0.2 mM CaCl2, 0.5 mM MnCl2, 0.01 mM CuCl2, 200 mM KCl, 15 mM NaCl, 25 mM HEPES, pH 7.4). The RNA was then refolded on ice for 5 min. Kinase reactions were initiated by adding thiophosphoryl donor(s) and moving the reaction mixtures to 32°C. For selection cycles 1–4, both 2.5 mM ATPγS and 2.5 mM GTPγS were present. The pool was then split, and cycles 5–10 were performed along separate paths in which only 2.5 mM ATPγS or 2.5 mM GTPγS was present. Aliquots of the Rnd6 products were pooled for a third path (denoted ‘mixed-donor’) in which both donors were present. Kinase reactions were stopped after 18 h (cycles 1–4) or 14 h (cycles 5–10) by addition of an equal volume of dye-free stop buffer (95% formamide, 15 mM EDTA). Samples were immediately loaded onto a trilayered 6% denaturing polyacrylamide gel with a 1–2 cm layer containing 50 µg/ml APM. This concentration of APM traps thiophosphorylated RNA at the non-APM/APM interface (42). Thiophosphorylated product was recovered from this layer by elution into dithiothreitol, as described (14). Purified RNAs were RT-PCR amplified for use in the next round of selection. Thiophosphorylated product was first visible at the APM interface in Rnd 7 for the A-specific and mixed-donor selections, and in Rnd 8 for the G-specific selection. Products of Rnd 10 were converted to double-stranded (ds) DNA and cloned into pUC18 for shotgun sequencing.
Kinetic analysis of kinase reactions
Internally radiolabeled transcripts were gel purified and refolded as above so that each reaction included 1–2 µM RNA (50 000–200 000 cpm). Reactions were initiated by addition of 2.5 mM donor (final concentration) for initial verification of activity or 1 mM donor for all other analyses, except where other concentrations are specified. Reaction mixtures were incubated at 32°C, then quenched at various times in stop buffer (95% formamide, 15 mM EDTA, xylene cyanol ff and bromophenol blue in traces). Products were separated on 8% denaturing trilayered APM gels. Autoradiographs were obtained with a FLA-5000 phosphorimager (FujiFilm) and analyzed with MultiGauge software. The fraction of the RNA converted to product at a given time [f(t)] was calculated by dividing the intensities of RNA retained at the APM interface into total for each lane. For kinetic analysis, the rate constants (kobs) and extrapolated plateaus (fmax) were obtained by fitting the data to a first-order rate equation using KaleidaGraph (Synergy Software):
Specificity of the phosphate donor
To determine donor specificity, product formation was measured for each ribozyme in the presence of 1 mM of each donor in separate reactions. Reactions were stopped after 8 h (GTPγS-dependent RNAs) or after 12 h (ATPγS-dependent RNAs) and analyzed as above. Analog competitions were performed in the presence of 1 mM cognate NTPγS plus 10 mM of each analog. Adenine stock solutions were prepared at low pH to promote solubility, and reactions were neutralized with NaOH.
Identification of thio-phosphorylation sites
Each ribozyme was labeled with 32P on its 5′ or 3′ terminus, gel purified, subjected to overnight self-thio-phosphorylation, and purified on an 8% APM-gel as above. Alkaline digestions of DTT-extracted, thio-phosphorylated products were performed in 2x Alk buffer (1x = 100 mM Na2CO3 pH 9.0, 2 mM EDTA), for 15 min at 90°C. Control RNAs were digested in 1x Alk buffer for 10 min. Ribonuclease T1 digestions under denaturing conditions (0.1 U/µl T1 in 0.025 M Na-citrate, 7 M urea, 10.5 mM EDTA pH 8.0, at 55°C for 10 min) served as size standards. Digestion products were separated on 10–20% denaturing polyacrylamide sequencing gels either with or without an APM layer.
Secondary structural analysis
Under native conditions at 32°C, 5′ radiolabeled RNA (50 000–200 000 cpm) was digested with ribonuclease T1 (Ambion, 0.005 U/µl for 2 min), or S1 nuclease (New England Biolabs, 4.75 U/µl for 10 min), or ribonuclease V1 (Ambion, 5 × 10−5 U/µl for 8 min). Size ladders for each RNA were produced via alkaline digestion and denaturing T1 digestions as above. All reactions were quenched with equal volumes of colorless gel loading buffer (10 M urea, 15 mM EDTA) and quickly cooled on dry ice plus ethanol. Products of digestions were separated on 8 M urea denaturing 15% polyacrylamide gels and analyzed as above. Secondary structures were calculated with the online web servers of the programs mFold (43) and MC-fold (44).
RESULTS
New populations of kinase ribozymes
A random-sequence RNA library was incubated overnight with ATPγS and/or GTPγS to allow for RNA-mediated thiophosphoryl transfer onto an internal 2′OH or terminal 2′/3′OH. Each RNA was 126 nt in length, with 70 random positions flanked by binding sites for amplification primers. Both donors were present during the first four selection cycles, after which the population was split into a G-, an A-specific path, and a third path in which both donors were present (‘mixed-donor’). In each selection cycle, thiophosphorylated RNA product was recovered from the organomercurial layer (N-acryloylamino-phenylmercuric chloride, or APM) of a tri-layer gel (14,42), amplified and transcribed for the next cycle of selection. Product accumulation above background was first detected in the 7th selection cycle and climbed to as high as 67% in the 10th cycle.
Approximately 20 sequences were obtained from each of the three selection paths (Supplementary Figure S1). Ribozymes from the A-specific path (K1–K20) clustered into six families of related sequences, while those obtained from the G-specific path (K21–K40) clustered into three families. Most (17 out of 20) of the ribozymes from the mixed-donor path (K41–K60) clustered with one of the A-specific families with 95–100% sequence identity, while three clones exactly matched the G1 family of G-specific ribozymes. To measure thiophosphoryl transfer activity of the individual selected ribozyme species, one representative from each sequence group was transcribed from PCR products, and these RNAs were incubated with ATPγS or GTPγS under the conditions of the selection. The products were again separated on a tri-layer APM gel and the fraction converted to product was plotted as a function of time (Figure 1A and B). Clone K19, the only representative of group A6, was inactive and was eliminated from subsequent analysis.
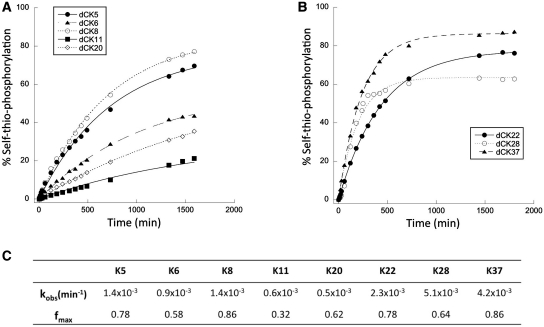
Figure 1.
Time-course of self-thio-phosphorylation reactions. Product formation was monitored under the selection conditions for individual kinase ribozyme clones that had been selected in the presence of ATPγS (A) or GTPγS (B) donors. Plotted data points are the averages of three independent replicas. Kinetic parameters of each ribozyme (C) were calculated by fitting the experimental data to a first-order exponential equation.
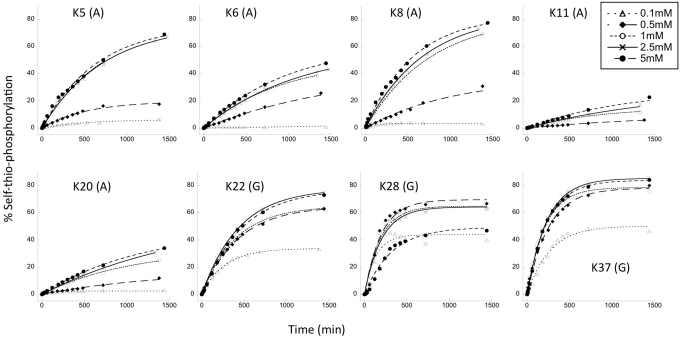
All eight active ribozymes followed first-order rate kinetics, with 30–90% conversion to product at the plateau. Individual rate constants (kobs) varied over a range of one order of magnitude (from 0.5 × 10−3 to 5.1 × 10−3 min−1) (Figure 1C), corresponding to a rate enhancement of approximately 104-fold relative to the rate of uncatalyzed hydrolysis of ATP (13,45). In general, ribozymes from the G-specific path (K22, K28, K37) were more active than those from the A-specific path, both in terms of kobs and fraction converted to product at the plateau. For the three ribozymes from the G-specific path, kobs values were all in the same order of magnitude (kobs = 4 × 10−3 min−1) and were essentially independent of donor concentration at all concentrations tested (KmGTPγS < 0.1 mM) (Figure 2, Supplementary Table S1). Each of the ribozymes from the A-specific path also gave similar kobs values (kobs = 0.9 × 10−3 min−1). These values were essentially independent of donor concentration at 0.5 mM and above, so we consider KmATPγS to be < 0.5 mM. (Net activity was too low to fit reliably at 0.1 mM for five of the six A-specific clones.) Subsequent experiments were performed at 1 mM ATPγS or GTPγS donor concentration, to ensure saturation of ribozyme active sites.
Figure 2.
Dependence of product formation on concentration of NTP donor. Identities of cognate ATPγS (A) and GTPγS (G) donors are given in parentheses. Concentrations are given in the legend in the upper right corner. Plotted data points are the averages of at least three replica, and were fitted with a first-order exponential equation.
Alternative donor nucleotide utilization
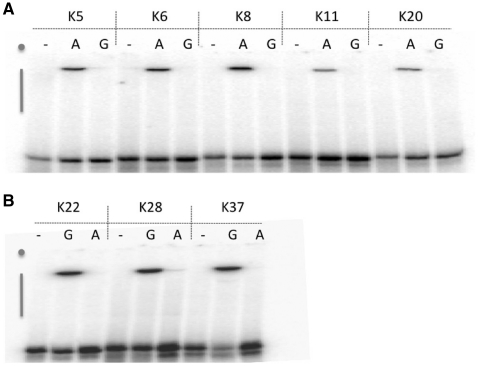
The A-specific and G-specific populations are derived from a common evolving population in which ATPγS and GTPγS donors were both present during the early selection cycles. Several ribozyme families were recovered from both the specific-donor and mixed-donor populations (in family A1, for example, six isolates are from the A-specific selection path and eight isolates are from the mixed-donor path). To determine whether this evolutionary history produced ribozymes with promiscuous substrate specificity, such as the ability to utilize any purine nucleotide as thiophosphoryl donor, activities were monitored for all eight active families in cross-donor assays, wherein molecules from the ATPγS selection were allowed to react with GTPγS, and molecules from the GTPγS selection were allowed to react with ATPγS. In every case, product formation was only observed with the cognate donor, and the signal in cross-donor assays was indistinguishable from background (Figure 3). Thus, recognition of the nucleobase portion of the donor nucleotides is essential for catalysis for each of the selected ribozymes.
Figure 3.
Donor specificity of ATPγS-dependent (A) and GTPγS-dependent (B) ribozyme clones. Tri-layered APM-PAGE of products of self-thio-phosphorylation in the presence of 1 mM ATPγS(A), 1 mM GTPγS(G) or no donor (–) above the figure. Gray dots indicate positions of wells, gray vertical bars denote positions of APM-containing polyacrylamide layers.
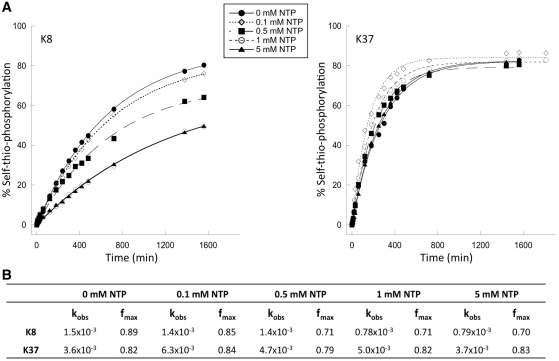
The gamma thio-substitution is required for efficient catalysis but not for substrate recognition
The most highly active clones from each group (K8 for the ATPγS group, and K37 for the GTPγS group) were subjected to self-thio-phosphorylation reactions in the presence of increasing concentrations of non-thio-substituted NTP as competitor. In each case, ribozyme kinetics were well behaved in the presence of the competitor (Figure 4A). Competition for access to the active site is expected to result in reduced rates of reaction as competitor concentration is increased without altering the plateau value at long reaction times. Independently, any phosphorylation by the natural (non-thio) nucleotide is expected to produce a poison product that will be unable to react with the thio-substituted nucleotide, thereby lowering the maximal amount of sulfur-containing product detected at the APM interface at long reaction times. For both ribozymes K8 and K37, kobs values for self-thio-phosphorylation in 1 mM competitor are half the values obtained when only the thio-substituted donor is present (kobsATPγS = 1.5 × 10−3min−1, kobsATPγS+ATP = 0.7 × 10−3 min−1 for K8; kobsGTPγS = 6.3 × 10−3 min−1, kobsGTPγS+GTP = 3.7 × 10−3 min−1 for K37). Thus, both ribozymes recognize the thio- and non-thio-substituted nucleotides with roughly equal affinity. The maximal amount of product formed at long times is essentially independent of 1 mM competitor for both ribozymes. The pattern was similar at lower competitor concentrations: reduced reaction rate with minimal effect on plateau values (Figure 4B). In separate reactions, several ribozymes became radiolabeled upon incubation with [γ-32P]ATP or [γ-32P]GTP (data not shown). However, the amount of product formed did not saturate within 24 h, and it was difficult to establish the fraction converted to product for kinetic analysis [in contrast to previous work with short oligonucleotide acceptor molecules (21)]. We conclude that for these two ribozymes, the sulfur atom on the gamma phosphate is required for efficient catalysis of thio-phosphoryl transfer but not for substrate recognition.
Figure 4.
Dependence of product formation on concentration of ATP and GTP competitors. (A) Time course of self-thio-phosphorylation reactions at 1 mM ATPγS (ribozyme K8) or GTPγS (ribozyme K37) in the presence of ATP or GTP competitors at various concentrations. (B) Observed rate constants (kobs, in units of min−1) and calculated plateau values (fmax) obtained by fitting the experimental data to a first-order exponential equation. Each data point is the average of at least three independent assays.
Donor nucleotide analog competition
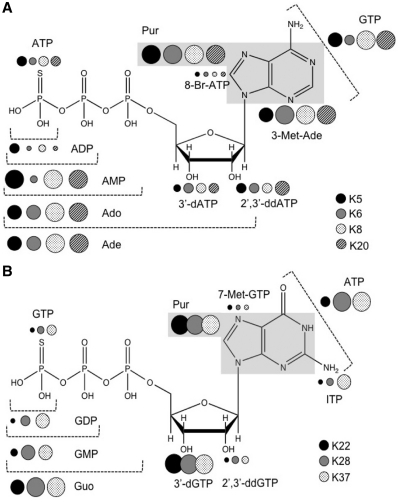
To obtain a more detailed view of substrate recognition by these ribozymes, reactions were monitored in the presence of 1 mM cognate donor and a panel of 10 mM analogs that served as non-reactive competitors. To avoid potential depletion of Mg2+ ions from chelation by the analogs, Mg2+ concentration was increased to 20 mM. To simplify analysis of multiple analogs for multiple RNAs, thiophosphorylation yields were determined at a single time point for each RNA, approximately half the time required to reach saturation in the absence of inhibitor. Low yield indicates competition with the cognate substrate, while high yield (little or no competition) implies that the chemical differences between substrate and analog prevents competition by interfering with recognition. Yield was normalized to the amount of product formed in the absence of competitor (Supplementary Figure S2). These results were mapped onto the structures of the cognate donors for four A-specific ribozymes (K5, K6, K8 and K20) and three G-specific ribozymes (K22, K28 and K37) (Figure 5) to define which moieties are required for donor nucleotide recognition. Large circles in the figure signify important contributions to substrate recognition from the indicated moiety.
Figure 5.
Summaries of nucleotide analogs competition assays. (A) Data for ATPγS analogs: ATP, ADP, AMP, adenosine (Ado), adenine (Ade), 3′-deoxy-ATP (3′-dATP), 2′,3′-dideoxy-ATP (2′,3′-ddATP), 8-bromo-ATP (8-Br-ATP), purine (gray box, Pur), 3-methyl adenine (3-Met-Ade) and GTP. (B) Data for GTPγS analogs: GTP, GDP, GMP, guanosine (Guo), 3′-deoxy-GTP (3′-dGTP), 2′,3′-dideoxy-GTP (2′,3′-ddGTP), purine (gray box, Pur), 7-methyl-GTP (7-Met-GTP), inosine triphosphate (ITP) and ATP. Guanine was not tested because of its low solubility at the pH of the reaction. Filled and shaded circles corresponding to each ribozyme are clustered next to the atoms or group of atoms whose roles are probed by the respective analogs. The sizes of the circles represent the percentage of self-thio-phosphorylation in five decreasing sizes relative to product formation for no-competitor reactions: >80%; 60–80%; 40–60%; 20–40%; <20%. Raw data (Supplementary Figure S2) were normalized to the control reaction, taken as 100% activity. Thus, large circles also indicate important contributions to substrate recognition. Specificities of ribozyme K11 were similar to those of the other ATPγS-dependent ribozymes; however, these data are omitted from Figure 5 because of the large error values relative to the measured yields (See Supplementary Figure S2).
Several trends are evident for each ribozyme and among subsets of selected ribozymes, with surprising convergence among the subsets. All eight ribozymes showed low reactivity (strong competition) when the γ-thio moiety was replaced with an oxygen in the analog. The sulfur atom is therefore not required for recognition, and the results obtained for K38 and K20 above can be extrapolated to all eight ribozymes tested here. All eight ribozymes were highly active in the presence of competitors that carry only the nucleoside or nucleobase components. Purine, adenine, 3-methyl-adenine and adenosine are all unable to compete for ATPγS, and neither purine nor guanosine competes with GTPγS. Competition progressively decreased as the gamma, beta and alpha phosphates were removed. Although the relative importance of the beta and gamma phosphates varied among the ribozymes, all eight ribozymes appear to use phosphate interactions as part of their overall recognition of their respective donors, and probably also as part of their reaction mechanism (see ‘Discussion’ section). The Watson–Crick faces of A and G, and the sugar edge of A are strong determinants of donor substrate recognition. This is evidenced by strong reactivity in the presence of GTP for the A group, or in the presence of ATP for the G group. Interestingly, inosine triphosphate strongly competes with GTPγS (see Supplementary Figure S2A), indicating that the exocyclic 2-amino group is not required for recognition by these three ribozymes. The 7-position (G group) and 8-position (A group) are not required for interaction by any of these ribozymes, as evidenced by strong competition from compounds with bulky moieties at these positions (7-methyl-GTP and 8-bromo-ATP). Finally, recognition requirements for the ribose hydroxyls are shared across all of the ribozymes from the A-specific path and across all of those from the G-specific path, although the two sets differ markedly in their dependences. The A-specific ribozymes are significantly inhibited by analogs that remove either the 2′OH or both the 2′ and 3′OH, indicating that those oxygens contribute little to donor substrate recognition. Although the G-specific ribozymes were also inhibited by 2′dGTP (no requirement for interaction with the 2′OH), they were not inhibited by 2′,3′-ddGTP (indicating a strong requirement for the 3′OH).
In addition to the inter- and intra-group similarities, some ribozymes displayed minor differences in their recognition patterns. For example, ribozymes K5 and K6 from the A-group are more dependent on the six-amino moiety than are ribozymes K8 and K20, as indicated by the weak competition from adenine in comparison with the complete lack of inhibition from adenine. Similarly, ribozyme K22 from the G-group is more readily inhibited by many of the analogs than are the other two ribozymes in this group. Overall, these results suggest a remarkable convergence of substrate recognition patterns both within and between ribozyme groups.
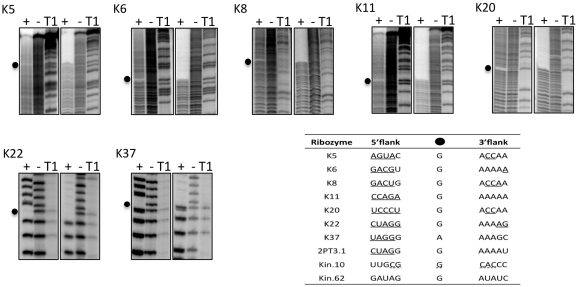
Identification of thio-phosphorylation sites
To map the precise sites of phosphorylation within each ribozyme, 5′-[32P]-labeled clones were self-thio-phosphorylated by incubation with the cognate NTPγS donor, followed by partial alkaline digestion. When the digested products were separated on denaturing sequencing gels, the modification site appears clearly as a gap in the digestion pattern (13,14). This approach not only identifies the nucleotide to which the thiophosphoryl group is attached, but also establishes the 2′OH as the point of attachment to the RNA chain, since this is the only stable attachment expected to prevent attack of the 2′OH on the adjacent phosphate. A single missing band in the ladder for the thiophosphorylated RNA identifies the modification site (Figure 6, left panels within each set of gels). As an independent strategy to identify modification sites, these same alkaline digestion products were separated on gels that contained an APM layer. For these analyses, all fragments containing the thiophosphoryl modification were retained at the top of the APM layer, while smaller, non-phosphorylated fragments formed a ladder well below the APM interface. The site of thiophosphorylation is identified as the first missing nucleotide above the ladder that passes through the APM layer (Figure 6, right panels within each set of gels). Both analytical procedures were also applied in separate reactions using 3′-radiolabeled RNA to ensure good resolution at both ends of the 126 nt RNAs (data not shown).
Figure 6.
Mapping thiophosphorylation sites. Autoradiography of alkaline digestion before (−) and after (+) self-thiophosphorylation of each 5′-radiolabeled clone. For each ribozyme, the same digestion products were run on normal (left three lanes) or on tri-layered APM (right three lanes) sequencing gels. Gaps in the digestion patterns (black dots) indicate thiophosphorylation sites. T1, size ladder generated by ribonuclease T1 digestion under denaturing conditions. Inset table shows the local sequence context of internal 2′ phosphorylation sites in kinase ribozymes selected in vitro. Ribozymes K5 through K37 are from this study. Ribozyme 2PT3.1 (29), ribozymes Kin.10 and Kin.62 (13) were isolated previously. Underlined nucleotides are involved in base pairing. Secondary structure of Kin.10 is speculative (dotted lines).
Both methods unambiguously identified a single thiophosphorylation site in each of seven ribozymes: K5, K6, K8, K11, K20, K22 and K37 are modified on nucleotides G71, G46, G45, G49, G40, G11 and A12, respectively. All of these sites are located on the 2′OH of purine residues, and all but one are on guanosines, independent of whether the ribozyme uses ATPγS or GTPγS as donor (see ‘Discussion’ section). The A-specific ribozymes self-modify within the original 70N random sequence between nucleotides 40 and 70, while the three G-specific ribozymes thio-phosphate one of two adjacent nucleotides within the 5′ primer-binding site. For the remaining ribozyme, K28, reaction products were consistently retained at the APM interface—indicating accumulation of self-thiophosphorylated product RNA—but there were no gaps in the digestion pattern. This behavior is consistent with the presence in the sample of mixed RNA populations carrying modifications at multiple sites, with no one site being fully occupied.
Secondary structural contexts of the phosphorylation sites
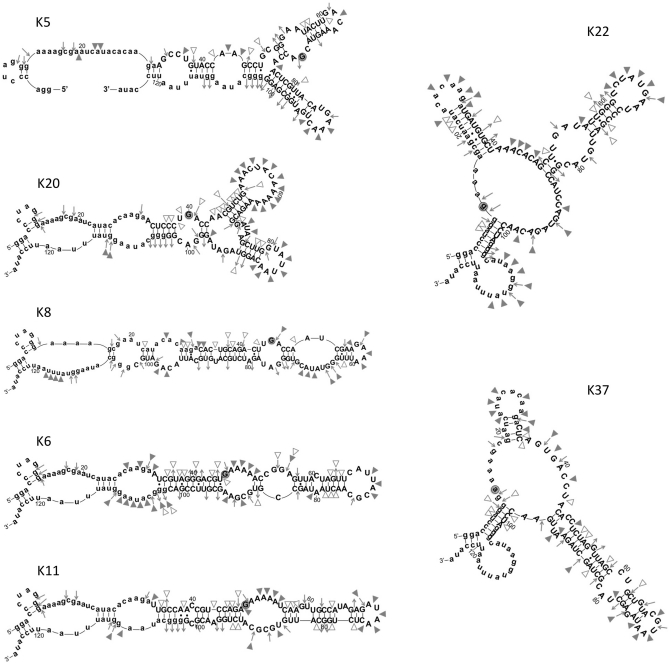
Secondary structures were determined for each of the ribozyme families whose modification sites were determined above. RNA (5′-end-labeled) was refolded in the kinase activity assay buffer and partially digested with RNAse T1 (cleaves after each unstructured G), V1 (cleaves ds RNA), or S1 (cleaves unstructured nucleotides). Cleavage sites were identified by gel electrophoresis (Supplemental Figure S3), and were used to identify probable secondary structures from among computational models generated by mFold (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) (43) and by MC-fold (http://www.major.iric.ca/MC-Fold/) (44) algorithms. The A-specific ribozymes fold into an elongated stem interrupted by multiple asymmetric internal loops (ribozymes K6, K8 and K11), or into branching structures with a central three-way junction (ribozymes K5 and K20). For the G-specific group, ribozymes K22 and K37 both fold into structures with three major helices that branch out from a central, three-way junction core with large unpaired segments (Figure 7). Both ribozymes K22 and K37 use the 5′ and 3′ primer-binding sites and a shared sequence from the original random nucleotides (AACCUA) to form one of the stems and joining strands.
Figure 7.
Predicted secondary structures of kinase ribozyme clones. Lower case letters refer to the original primer binding sites. Symbols indicated cleavage sites in 1× phosphorylation buffer: filled triangles, S1 nuclease (cleaves unstructured RNA); open triangles, V1 nuclease (cleaves double-stranded RNA); inward-pointing arrows, ribonuclease T1 (cleaves after unstructured G residues); outward-pointing arrows, G residues that are not cleaved by ribonuclease T1 (See Supplementary Figure S3 for an example of the structural probing primary data).
The modification sites identified above are all located on unpaired nucleotides within internal loops. Three structural contexts are apparent. For ribozymes K5, K8 and K20, the modified nucleotide is in the middle of a 3 nt YGA motif within a symmetrical, internal loop. The unmodified strand of the internal loop is the sequence GAN for all three of these ribozymes. For ribozymes K6 and K11, the modification sites are within an A-rich strand on the G residue immediately adjacent to a base-paired stem, and the unlabeled strand is of the sequence UGCG(C/A)A. This motif is identical to the active site of the 2PT3.1 self-kinasing ribozyme identified previously (14). Unlike ribozymes K6 and K11, ribozyme 2PT3.2 modifies a guanosine in the second position of the purine-rich strand rather than the position immediately adjacent to the stem. The purine-rich strand of the active site in 2PT3.1 was derived from the fixed, primer-binding site, while in ribozymes K6 and K11 these segments are derived completely from the original 70N random segments, further indicating their independent evolutionary origins. For the two G-specific ribozymes, the site of modification is a guanosine immediately adjacent to (ribozyme K22), or an adenosine one position removed from (ribozyme K37), a base-paired stem. Both modification sites are within the three-way junction and are across from two or more adenosines in the AACCUA sequence motif shared between these isolates. Thus, the independent structures formed by the seven A- and G-specific ribozymes described here converge on a restricted set of three secondary structures to achieve thiophosphoryl transfer onto internal 2′OH.
DISCUSSION
This work describes kinase activity and substrate recognition for eight new families of kinase ribozymes. Ribozymes that utilize GTPγS (K22, K28, K37) are slightly more active than their ATPγS counterparts with respect to the parameters that determine selection fitness (rate, donor substrate affinity and fraction folded into a productive conformation). The apparent Km values (Km <0.1 to <0.5 mM) of all the selected species are well below the concentration of donor nucleotides that were available during the selection (2.5 mM), and the apparent first-order rate constants (kobs = 1 to 4 × 10−3 min−1) correspond to 1–5 reaction half-lives during the overnight incubations. Thus, the selection yielded ribozymes for which the selection conditions were highly permissive both in terms of effecting chemistry and substrate recognition. These apparent Km and kcat values are comparable to those obtained previously for unoptimized kinase (deoxy)ribozymes (13–15,17).
Ten internal thiophosphorylation sites have now been mapped in kinase ribozymes derived from random- or nearly random-sequence populations [seven in this work and three mapped previously (13,14)]. All 10 are on purines, and 9 of 10 are on guanosines, irrespective of whether the ribozymes utilized ATPγS or GTPγS as thiophosphoryl donor (see inset in Figure 6). Intrinsic nucleophilicity and positioning effects may contribute to this non-random regioselectivity. A recent study of 2′OH alkylation susceptibilities in denatured RNA found that the 2′OH of A and G are indistinguishably reactive, and that they are 1.4–1.7 times more reactive than the pyrimidines hydroxyls (46). Ribose 2′OH pKa values measured directly by proton chemical shift titration (47,48) each give slightly different pKa values, but the trend is that the 2′OH of A is a few tenths of a pH unit more acidic than G, followed closely by the pyrimidines. However, indirect measurements of pKa using transesterification kinetics (49,50) indicated that the identities of the nucleotide bases that flank the target RNA linkage have a negligible effect on the pKa of the nucleophilic 2′OH (49). Thus, while intrinsic chemical reactivity modestly favors purines, it does not fully explain the preference for guanosine.
We speculate that substrate orientation plays an important role in determining the targeted nucleotide. Nucleophilic displacement at phosphates is optimal when nucleophile and leaving group are oriented 180° relative to the phosphorous (tau angle) and the nucleophile is at a distance of 3 Å. The resulting ‘in-line fitness’ falls off approximately as the squares of the distance and of cos(τ) (51). The multiple interaction opportunities presented by the guanine nucleobase (especially via hydrogen bonding and aromatic stacking) may simplify the sequence requirements for the formation of a ribozyme acceptor pocket that orients the 2′OH for productive chemistry. It is important to note that internal esterification on cytidine (52) and uridine (53) has been noted previously for (amino)acyl transfer from AMP- or CoA-activated acyl groups, demonstrating that regioselectivity for the guanosine 2′OH is not universal. Furthermore, reselection of thiophosphoryl transfer activity from a partially mutagenized acyltransfer ribozyme yielded internal phosphorylation at all four nucleotides in roughly equal numbers (A:C:G:U = 1:1:2:1) (15) (these results are not included in the inset in Figure 6 because the starting library was heavily constrained by the original acyltransferase ribozyme sequence from which those output kinase ribozymes were selected). Nevertheless, the combination of subtle preferences, such as purine nucleophilic chemistry and acceptor orientation, may have been amplified through multiple cycles of selection to yield the observed overabundance of guanosines as thiophosphoryl acceptors.
The eight ribozymes analyzed here showed a remarkable convergence of apparent requirements for donor substrate recognition, as defined through analog competition assays and alternative substrate utilization assays. In all cases, the nucleobase appears to be critical to recognition, especially the Watson–Crick faces of G and A and the sugar face of A. This observation can be explained, in part, by the fact that the selections started with both ATPγS and GTPγS present, thereby eliminating from the population ATPγS-dependent ribozymes that are strongly inhibited by GTP and GTPγS-dependent ribozymes that are inhibited by ATP. Analogs with bulky substitutions on the Hoogsteen face (7-methyl-GTP and 8-bromo-ATP) compete with the cognate substrates, indicating a lack of steric constraints at these positions. The G-specific ribozymes appear to hydrogen bond to the ribose 3′OH, while the A-specific ribozymes are insensitive to ribose alterations. While DNAzymes that phosphorylate 5′OH groups had previously been shown to differentiate among the major nucleotide triphosphates (17), the present work provides the most detailed view to date regarding the individual moieties responsible for donor nucleotide recognition. The features of nucleotide recognition reported here parallel those observed previously for aptamers selected to bind nucleotides (22,24,35).
Importantly, all of these ribozymes also show evidence of interacting with the alpha, beta and/or gamma phosphates. Removing the phosphates progressively reduces competition for the active site. Such interactions may help position the gamma phosphorous and beta/gamma bridging oxygen for in-line, nucleophilic displacement by the attacking acceptor oxygen. While several naturally-occurring riboswitches recognize phosphates in their target metabolites (54–58), phosphate recognition is often accompanied by more complex RNA structures. Phosphate recognition is very rare among aptamers selected from random-sequence libraries, such as the 3′ phosphate requirement for CoA aptamers 80PSA21 and 70PSA17 (29) and modest discrimination between cAMP and adenosine (32), or between FMN and riboflavin (33). Most aptamers selected in vitro do not discriminate between phosphorylated and non-phosphorylated forms of their molecular targets (22,23,25–28,30,31,35,59,60). Thus, the requirements for donor substrate utilization impose additional selection pressures on evolving ribozymes beyond those encountered by aptamers that are selected merely for binding to their ligands by extending recognition to include one or more phosphates.
From this and previous work, it is clear that random sequence space is populated with a large number of structured RNAs that catalyze self-thiophosphorylation of terminal or backbone hydroxyls. (Thio)Phosphoryl transfer onto diffusible metabolites is anticipated to be more challenging catalysis due to the need to orient the acceptor. Further exploration of ribozyme capabilities will establish whether metabolically relevant kinase ribozymes could have arisen readily in an RNA world.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Aeronautics and Space Administration Exobiology program (grant NAG5-12360 to D.H.B.); by a Life Sciences postdoctoral fellowship from the University of Missouri to E.B. Funding for open access charge: University of Missouri.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank undergraduate researchers James Patterson and John Zaborske for technical assistance and stimulating discussions early in the project, and Dr Mark Ditzler, Dr Margaret Lange and two anonymous reviewers for critical reading of the manuscript.
REFERENCES
- 1.Burke DH. Ribozyme-catalyzed genetics. In: Ribas de Pouplana L, editor. The Genetic Code and the Origin of Life. © 2004 Eurekah.com and Kluwer Academic/Plenum Publishers; 2004. pp. 48–74. [Google Scholar]
- 2.Nissen P, Hansen J, Ban N, Moore P, Steitz T. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 3.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 4.Dambach MD, Winkler WC. Expanding roles for metabolite-sensing regulatory RNAs. Curr. Opin. Microbiol. 2009;12:161–169. doi: 10.1016/j.mib.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 6.Wiegand TW, Janssen RC, Eaton BE. Selection of RNA amide synthases. Chem. Biol. 1997;4:675–683. doi: 10.1016/s1074-5521(97)90223-4. [DOI] [PubMed] [Google Scholar]
- 7.Tarasow TM, Tarasow SL, Eaton BE. RNA-catalyzed carbon-carbon bond formation. Nature. 1997;389:54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- 8.Wilson C, Szostak JW. In vitro evolution of a self-alkylating ribozyme. Nature. 1995;374:777–782. doi: 10.1038/374777a0. [DOI] [PubMed] [Google Scholar]
- 9.Lohse PA, Szostak JW. Ribozyme-catalyzed amino-acid transfer reactions. Nature. 1996;381:442–444. doi: 10.1038/381442a0. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 11.Ekland EH, Bartel DP. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature. 1996;382:373–376. doi: 10.1038/382373a0. [DOI] [PubMed] [Google Scholar]
- 12.Illangasekare M, Sanchez G, Nickels T, Yarus M. Aminoacyl-RNA synthesis catalyzed by an RNA. Science. 1995;267:643–647. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]
- 13.Lorsch JR, Szostak JW. In vitro evolution of new ribozymes with polynucleotide kinase activity. Nature. 1994;371:31–36. doi: 10.1038/371031a0. [DOI] [PubMed] [Google Scholar]
- 14.Saran D, Nickens D, Burke D. A trans acting ribozyme that phosphorylates exogenous RNA. Biochemistry. 2005;44:15007–15016. doi: 10.1021/bi051086h. [DOI] [PubMed] [Google Scholar]
- 15.Curtis E, Bartel D. New catalytic structures from an existing ribozyme. Nat. Struct. Mol. Biol. 2005;12:994–1000. doi: 10.1038/nsmb1003. [DOI] [PubMed] [Google Scholar]
- 16.Saran D, Held D, Burke D. Multiple-turnover thio-ATP hydrolase and phospho-enzyme intermediate formation activities catalyzed by an RNA enzyme. Nucleic Acids Res. 2006;34:3201–3208. doi: 10.1093/nar/gkl413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Breaker R. Phosphorylating DNA with DNA. Proc. Natl Acad. Sci. USA. 1999;16:2746–2751. doi: 10.1073/pnas.96.6.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Billen LP, Li Y. Sequence diversity, metal specificity, and catalytic proficiency of metal-dependent phosphorylating DNA enzymes. Chem. Biol. 2002;9:507–517. doi: 10.1016/s1074-5521(02)00127-8. [DOI] [PubMed] [Google Scholar]
- 19.Achenbach JC, Jeffries GA, McManus SA, Billen LP, Li Y. Secondary-structure characterization of two proficient kinase deoxyribozymes. Biochemistry. 2005;44:3765–3744. doi: 10.1021/bi0483054. [DOI] [PubMed] [Google Scholar]
- 20.McManus SA, Li Y. Multiple occurrences of an efficient self-phosphorylating deoxyribozyme motif. Biochemistry. 2007;46:2198–2204. doi: 10.1021/bi061613c. [DOI] [PubMed] [Google Scholar]
- 21.Lorsch JR, Szostak JW. Kinetic and thermodynamic characterization of the reaction catalyzed by a polynucleotide kinase ribozyme. Biochemistry. 1995;34:15315–15327. doi: 10.1021/bi00046a041. [DOI] [PubMed] [Google Scholar]
- 22.Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 23.Burgstaller P, Famulok M. Isolation of RNA aptamers for biological cofactors by in vitro selection. Angew. Chem., Int. Ed. Engl. 1994;33:1084–1087. [Google Scholar]
- 24.Burke DH, Gold L. RNA aptamers to the adenosine moiety of S-adenosyl methionine: Structural inferences from variations on a theme and the reproducibility of SELEX. Nucleic Acids Res. 1997;25:2020–2024. doi: 10.1093/nar/25.10.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 26.Lato SM, Ozerova NDS, He K, Sergueeva Z, Shaw BR, Burke DH. Boronated RNA aptamers to ATP. Nucleic Acids Res. 2002;30:1401–1407. doi: 10.1093/nar/30.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis J, Szostak J. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl Acad. Sci. USA. 2002;99:11616–11621. doi: 10.1073/pnas.182095699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke DH, Hoffman DC. A novel acidophilic RNA motif that recognizes coenzyme A. Biochemistry. 1998;37:4653–4663. doi: 10.1021/bi972877p. [DOI] [PubMed] [Google Scholar]
- 29.Saran D, Frank J, Burke DH. The tyranny of adenosine recognition among RNA aptamers to coenzyme A. BMC Evol. Biol. 2003;3:26. doi: 10.1186/1471-2148-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roychowdhury-Saha M, Lato SM, Shank ED, Burke DH. RNA aptamers that bind FAD. Biochemistry. 2002;41:2492–2499. doi: 10.1021/bi015719d. [DOI] [PubMed] [Google Scholar]
- 31.Lauhon CT, Szostak JW. RNA aptamers that bind flavin and nicotinamide redox cofactors. J. Am. Chem. Soc. 1995;117:1246–1257. doi: 10.1021/ja00109a008. [DOI] [PubMed] [Google Scholar]
- 32.Koizumi M, Breaker R. Molecular recognition of cAMP by an RNA aptamer. Biochemistry. 2000;39:8983–8992. doi: 10.1021/bi000149n. [DOI] [PubMed] [Google Scholar]
- 33.Anderson PC, Mecozzi S. Identification of a 14mer RNA that recognizes and binds flavin mononucleotide with high affinity. Nucleic Acids Res. 2005;33:6992–6999. doi: 10.1093/nar/gki992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dieckmann T, Suzuki E, Nakamura GK, Feigon J. Solution structure of an ATP-binding RNA aptamer reveals a novel fold. RNA. 1996;2:628–640. [PMC free article] [PubMed] [Google Scholar]
- 35.Carothers JM, Davis JH, Chou JJ, Szostak JW. Solution structure of an informationally complex high-affinity RNA aptamer to GTP. RNA. 2006;12:567–579. doi: 10.1261/rna.2251306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villa A, Wöhnert J, Stock G. Molecular dynamics simulation study of the binding of purine bases to the aptamer domain of the guanine sensing riboswitch. Nucleic Acids Res. 2009;37:4774–4786. doi: 10.1093/nar/gkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan P, Suri AK, Fiala R, Live D, Patel DJ. Molecular recognition in the FMN-RNA aptamer complex. J. Mol. Biol. 1996;258:480–500. doi: 10.1006/jmbi.1996.0263. [DOI] [PubMed] [Google Scholar]
- 38.Jiang F, Kumar RA, Jones RA, Patel DJ. Structural basis of RNA folding and recognition in an AMP-RNA aptamer complex. Nature. 1996;382:183–186. doi: 10.1038/382183a0. [DOI] [PubMed] [Google Scholar]
- 39.Levy M, Ellington AD. RNA world: catalysis abets binding, but not vice versa. Curr. Biol. 2001;11:R665–R667. doi: 10.1016/s0960-9822(01)00388-8. [DOI] [PubMed] [Google Scholar]
- 40.Cho B, Burke D. Topological rearrangement yields structural stabilization and interhelical distance constraints in the Kin.46 self-phosphorylating ribozyme. RNA. 2006;12:2118–2125. doi: 10.1261/rna.173506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Igloi G. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry. 1988;27:3842–3849. doi: 10.1021/bi00410a048. [DOI] [PubMed] [Google Scholar]
- 42.Rhee S, Burke D. Tris(2-carboxyethyl)phosphine stabilization of RNA: comparison with dithiothreitol for use with nucleic acid and thiophosphoryl chemistry. Anal. Biochem. 2004;325:137–143. doi: 10.1016/j.ab.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 45.Ramirez F, Marecek JF, Szamosi J. Magnesium and calcium ion effects on hydrolysis rates of adenosine 5′-triphosphate. J. Org. Chem. 1980;45:4748–4752. [Google Scholar]
- 46.Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15:1314–1321. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velikyan I, Acharya S, Trifonova A, Foldesi A, Chattopadhyaya J. The pKa’s of 2′-hydroxyl groups in nucleosides and nucleotides. J. Am. Chem. Soc. 2001;123:2983–2984. doi: 10.1021/ja0036312. [DOI] [PubMed] [Google Scholar]
- 48.Izatt RM, Hansen LD, Rytting JH, Christensen JJ. Proton ionization from adenosine. J. Am. Chem. Soc. 1965;87:2760–2761. doi: 10.1021/ja01090a044. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Breaker R. Kinetics of specific base catalysis of RNA degradation by transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999;121:5364–5372. [Google Scholar]
- 50.Jarvinen P, Lonnberg H, Oivanen M. Interconversion and phosphodiester hydrolysis of 2′,5′ and 3′,5′-dinucleoside mono- phosphates: kinetics and mechanisms. J. Org. Chem. 1991;56:5396–5401. [Google Scholar]
- 51.Soukup G, Breaker R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenne A, Famulok M. A novel ribozyme with ester transferase activity. Chem. Biol. 1998;5:23–34. doi: 10.1016/s1074-5521(98)90084-9. [DOI] [PubMed] [Google Scholar]
- 53.Li N, Huang F. Ribozyme-catalyzed aminoacylation from CoA thioesters. Biochemistry. 2005;44:4582–4590. doi: 10.1021/bi047576b. [DOI] [PubMed] [Google Scholar]
- 54.Jansen JA, McCarthy TJ, Soukup GA, Soukup JK. Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme. Nat. Struct. Mol. Biol. 2006;13:517–523. doi: 10.1038/nsmb1094. [DOI] [PubMed] [Google Scholar]
- 55.Noeske J, Richter C, Stirnal E, Schwalbe H, Wohnert J. Phosphate-group recognition by the aptamer domain of the thiamine pyrophosphate sensing riboswitch. ChemBioChem. 2006;7:1451–1456. doi: 10.1002/cbic.200600151. [DOI] [PubMed] [Google Scholar]
- 56.Thore S, Frick C, Ban N. Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J. Am. Chem. Soc. 2008;130:8116–8117. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- 57.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burke DH, Scates LA, Andrews K, Gold L. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J. Mol. Biol. 1996;264:650–666. doi: 10.1006/jmbi.1996.0667. [DOI] [PubMed] [Google Scholar]
- 60.Carothers J, Oestreich S, Davis J, Szostak J. Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 2004;126:5130–5137. doi: 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.