Abstract
Homologous recombination is a fundamental process enabling the repair of double-strand breaks with a high degree of fidelity. In prokaryotes, it is carried out by RecA nucleofilaments formed on single-stranded DNA (ssDNA). These filaments incorporate genomic sequences that are homologous to the ssDNA and exchange the homologous strands. Due to the highly dynamic character of this process and its rapid propagation along the filament, the sequence recognition and strand exchange mechanism remains unknown at the structural level. The recently published structure of the RecA/DNA filament active for recombination (Chen et al., Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structure, Nature 2008, 453, 489) provides a starting point for new exploration of the system. Here, we investigate the possible geometries of association of the early encounter complex between RecA/ssDNA filament and double-stranded DNA (dsDNA). Due to the huge size of the system and its dense packing, we use a reduced representation for protein and DNA together with state-of-the-art molecular modeling methods, including systematic docking and virtual reality simulations. The results indicate that it is possible for the double-stranded DNA to access the RecA-bound ssDNA while initially retaining its Watson–Crick pairing. They emphasize the importance of RecA L2 loop mobility for both recognition and strand exchange.
INTRODUCTION
Homologous recombination (HR) is a fundamental process allowing the repair of double strand breaks with a high degree of fidelity (1–3). This process exists throughout the whole realm of life and involves recombinase proteins such as RecA for prokaryotes and Rad51 for eukaryotes. When a double-strand break appears in DNA, one of the broken strands is degraded (4), while the other gets covered by monomers of a recombinase, organized as a right-handed helical filament. The central event in HR involves sequence recognition between the single-stranded DNA (ssDNA) on which the recombinase has polymerized and double stranded DNA stretches from the genome (dsDNA). Successful recognition results in the exchange of homologous ssDNA and dsDNA strands (Supplementary Data 0).
In order to understand the mechanism of recognition and strand exchange, it is necessary to gain structural information on the recognition intermediates. However, these intermediates are very short lived and therefore are difficult to access using structural biophysics methods. In spite of the remarkable work that recently led to the resolution of RecA filament crystal structures in complex with DNA (5), we only dispose of snapshots of the filament, prior or posterior to sequence recognition. Still, these structures are precious starting points to investigate the initial geometries of association between RecA/ssDNA filaments and dsDNA. In the present study, we explore the possible geometries of association at the very initial phase of recognition, when dsDNA first interacts with the filament interior in search for homology. Our aim is to provide structural insight in the longstanding question of the HR mechanism.
This study takes advantage of our recently developed coarse - grained model of DNA, devoted to protein/DNA docking in reduced representation (6). The minimal filament size for a meaningful (free from end effects) study of DNA association to the nucleofilament is of the order of one helical turn (around six RecA monomers). The crystallographic RecA/DNA filaments solved by Chen et al. (5) contain from five to six RecA monomers, and are therefore adapted to the study. The dimension of the structure identified by the PDB code 3CMX (RecA/DNA with five RecA monomers) is about 150 Å height versus 80 Å diameter, for a total of about 12 500 atoms if only the single strand initially bound to RecA is considered (chains A and B). In addition, the incoming DNA fragment must be sufficiently long to interact with every part of the filament. Consequently, the huge size of the system makes it difficult to exhaustively investigate possible DNA association geometries at atomic resolution. For this reason, we chose to initiate the study at low resolution and in rigid body mode using the docking program ATTRACT (7,8), which systematically explores all possible arrangements of macromolecules, either proteins or DNA, via series of energy minimizations starting from multiple positions and orientations. We are confident in relying on the ATTRACT protocol and force field since this approach generated reliable predictions for six protein/DNA complexes that cover a large range of diversity, in terms of mode of association and DNA deformation (6).
Due to their high degree of flexibility, the structure of the L2 loops that line the interior of the nucleoprotein filament is investigated in a second stage of our study, in relation with dsDNA positioning. These long loops (20 amino acids) are known to be disordered in the absence of DNA except under specific crystallization conditions, such as those used to obtain the structure of Mycobacterium tuberculosis RecA (MtRecA, PDB code 1MO4) (9). Indeed, as anticipated from previous modeling work (10,11), L2 loops can largely span the central space of the filament interior. In the RecA filament structure obtained in the presence of DNA, they cross the filament axis to fold upon the RecA-bound DNA, while in the case of MtRecA they remain closer to the protein surface. It is thus expected that L2 will interfere in some way with the incoming dsDNA, either by favoring or by hindering its approach. Here, we investigate the degree of L2 loop distortion that would be necessary to permit an incoming dsDNA to scan the sequence of RecA-bound ssDNA, using low-resolution interactive simulations. This new technique, based on extensions of the MDDriver library (12–14) and the MyPal application (15) with the benefit of haptic devices, appears as a powerful tool enabling the user to simultaneously impose, control and visualize both L2 internal deformations and dsDNA position/orientation with respect to the filament.
The possible distortion induced on incoming dsDNA during association is investigated by successively docking oligonucleotides of identical sequence but presenting two different structures, a canonical B-DNA structure and a curved structure. The latter choice merits some comments. We have previously pointed out the structural similarities between the stretched and unwound DNA form found in recombinase filaments and the local deformations induced in DNA by architectural proteins (10). These proteins generally bind the DNA minor groove and bend the double helix by amplitudes that reach 90° [for TATA-box bound to TBP (16)], without altering its internal Watson–Crick connectivity. Such dramatic distortion has been described as resulting from the junction between the physiological B-form DNA and a distorted section in contact with the protein (17). This section called AT-form, unwound with widened minor groove, is characterized by the formation of important kinks at the frontier with B-DNA. Both RecA-bound (10,18) and TBP-bound DNA forms (17,19) have been simulated using exactly the same modeling process, i.e. by exerting a stretching restraint on the 3′3′ extremities of both antiparallel DNA strands. In the case of TBP-bound DNA, applying the restraints to the DNA section contacting the protein correctly reproduced the deformation, including the flanking kinks. Alternatively, the restraints were applied to the whole length of the DNA molecule for RecA-bound DNA. Note that the model proposed in this last case, >10 years ago, remarkably coincides with the recently solved RecA-bound DNA crystal structure (20).
In this context, a plausible hypothesis is that during exploratory contact between dsDNA and the nucleofilament, the RecA-induced dsDNA distortion would be concentrated on the section directly in contact with RecA/ssDNA, with kinks resulting at the junction between this section and the free DNA. For this reason, the second DNA structure that we dock to the RecA/ssDNA nucleofilament presents the characteristic deformations of the TBP-bound TATA-box. Our investigations bear on the practical feasibility for such a structure to penetrate the filament groove and probe the ssDNA sequence, together with the potential consequences in terms of sequence recognition.
MATERIALS AND METHODS
Docking simulations
The system formed by RecA/ssDNA and dsDNA was investigated at low resolution, using the docking program ATTRACT (7,8) and our recently developed DNA coarse-grained model, enabling systematic docking of protein/DNA systems (6).
The coarse-grained representation and force field used in ATTRACT have been fully described in references (6,7). Both protein and DNA coarse-grained models are characterized by a reduction of system size by a factor 3–4 in terms of heavy atoms per bead. The associated force field only contains terms for the interaction energy between partners, composed of a soft Lennard–Jones term and an electrostatic potential term. This level of reduction allows conservation of the main features of the surface of each partner. We have shown that the model ensures the stability of protein/DNA complexes and permits to identify the correct geometry of various types of such complexes. Whereas the present version of ATTRACT does not account for DNA flexibility, the low resolution produces reliable results even when using DNA structures that moderately differ from their structure in the complex, tolerating up to 20° bending deformations (6).
The ATTRACT program allows systematic search of the relative orientations and translations of two partners by performing a series of multiple energy minimizations with respect to translation and rotation degrees of freedom, starting from initial positions regularly distributed around the receptor (arbitrarily defined as the biggest of the two partners) and hundreds of starting orientations for each initial position, here amounting to 400. The resulting geometries are classified based on surface and electrostatic complementarity using the simplified interaction energy terms described above. We have previously shown the good performance of the method in terms of conformational space exploration. In a previous protein–DNA study on several test cases, the near-native docking solution was always found as best ranked complex several times, indicating that the number of starting geometries is sufficient. In addition, docking simulations of systems with palindromic DNA predicted two clusters of correct geometries with identical sizes, both of them ranked first in energy, where the DNA was oriented either in the crystal orientation or upside down (6). The density of start configurations (position and orientation) used in the present study correspond to the optimal conditions used in that previous study.
Docking dsDNA to the RecA/ssDNA filament required 145 200 minimization runs starting from 363 different positions, constructed as follows. Points distant by 10 Å from each other were generated around the filament at a distance of 28 Å from the surface. From this ensemble, points situated within a crown, 98 Å height, centered around the filament axis (153 Å long) were conserved. Eliminating the points at both extremities of the filament avoided to search non-relevant protein regions for dsDNA interaction, which have moreover been engineered in the crystal structure of the RecA/ssDNA complex (5).
Using ATTRACT, a 24 bp dsDNA could be docked on RecA/ssDNA within a few hours.
RecA nucleofilament
The nucleofilament was taken in its extended form that is active for recombination (PDB code 3CMX) (5). The structure contains five RecA monomers bound to DNA. From the nucleic part, we only kept the DNA strand of sequence (dT)13 bound to the first binding site. The complementary strand is only used as a reference in the discussion of Figure 3. In a first series of docking simulations, the four L2 loops present in the structure (residues L2_0: 193 to 212, L2_1: 1193 to 1212, L2_2: 2193 to 2212, L2_3: 3193 to 3212; loop L2 from the fifth RecA monomer is disordered) were deleted to investigate how close the intact dsDNA can possibly approach the ssDNA buried within the filament. The protein and nucleic parts of the filament were separately translated into a reduced representation using the PTools library (8), then merged back into the receptor file. In the process, the total number of particles in the system was reduced from 12 508 Atoms to 3742 beads.
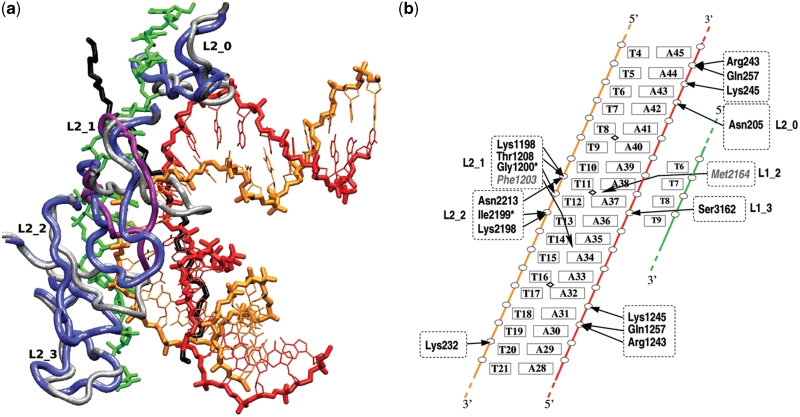
Figure 3.
Two views of the atomic model for the encounter complex. (a) The dsDNA (same color code as in Figure 2) contacts the ssDNA (green) via its minor groove. The starting (from 3CMX) and modeled L2 loop structures are, respectively, in gray and blue (tube representation). Loop L2_1 from MtRecA is in magenta. The protein scaffold is not represented. The black segment corresponds to the phosphodiester backbone of the complementary strand in the 3CMX crystal structure. This segment is not part of the model but has been added to the representation for comparison, since it coincides with the dsDNA complementary strand at the level of loop L2_1. (b) Schematic description of the dsDNA/ssDNA/L2 interactions. The color codes for the three DNA strands correspond to those in (a). Except for residues Phe1203 and Met2164, arrows indicate salt bridges or hydrogen bonds between protein residues and DNA phosphate groups (white circles). In two cases indicated by asterisk, the residues interact via their main chain NH group. The residues in gray and italics are positioned in the major groove for Phe1203 and the minor groove for Met2164. Diamond signs indicate the kink locations in DSC structure.
This reduced representation was used both for systematic rigid body docking and for flexible interactive molecular simulations.
DNA structures
Two distinct dsDNA structures were docked to the filament, a B-DNA form called DSB and a curved form called DSC. The 24 bp long DSB structure was constructed using the program JUMNA (21,22) with standard helical parameters. The sequence was taken homologous to that of the RecA-bound ssDNA, i.e. d(T24.A24). The DSC structure was constructed from the crystal structure of the TBP-bound TATA-box of sequence d(GTATATAAAACG) (PDB code 1YTB), extended by 6 bp at both extremities. To this end, the helical parameters of 1YTB with respect to a best fitting axis kinked at two positions (at base pair steps T2pA3 and A10pC11) were extracted using the program Curves (23,24). Standard B-DNA parameters for 6 bp were then appended at each extremity of the parameter file, which was finally applied to the d(T24.A24) sequence using JUMNA. The final structure is a 24-bp long oligonucleotide presenting the 1YTB distortion at its center (represented in Figures 2b and 3). For DSB as well as for DSC, the nucleotides are numbered from the 5′ to the 3′-side, from 1 to 24 for the d(T) strand (also called homologous strand) and from 25 to 48 for the complementary d(A) strand.
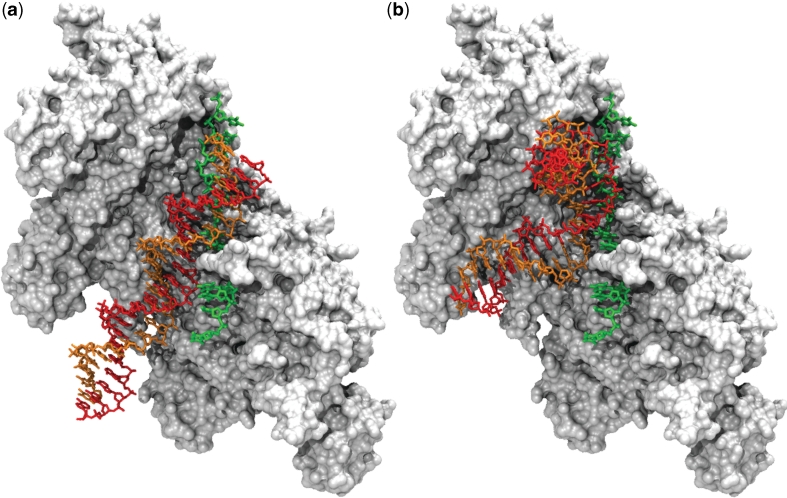
Figure 2.
Result of a docking simulation between a double-stranded B-DNA (DSB) (a) or a curved DNA (DSC) (b) and the RecA/ssDNA filament. The dsDNA is in red for the complementary strand and orange for the strand homologous to the ssDNA. The ssDNA is in green and the surface representation of the protein is white/gray. For a better understanding, the partners of association have been represented at atomic resolution. To this aim, the initial structures (before translation into coarse-grained representation) of DSB (a) or DSC (b) were submitted to exactly the same geometric transformations as those resulting from the docking simulation (8).
Analysis
Analysis of the exploratory simulations of the encounter complex was based not only on the interaction energy used as an indicator of the best geometries of association in the ATTRACT protocol (6), but also on additional criteria specific to the system, for the following reasons. First, the investigated complex is transient; therefore, it is expected to show little stabilization with respect to alternative geometries of association. Although our low-resolution force field is sufficient to clearly discriminate the correct geometry of association for stable complexes, it was not designed to manage such delicate energy balances. In addition, due to the absence of the L2 loops, we expect the energy criterion to be inadequate since the system lacks not only the loop charges, but also putative stabilizing van der Waals interactions between the L2 loops and the incoming dsDNA. Such interactions are expected due to the crowded environment within the filament and from comparison to RecA-bound DNA in both RecA/ssDNA and RecA/dsDNA crystal structures, where dsDNA is stabilized by L2 loops (5). A last reason is that ions are not included in the simulation, while multivalent ions such as Mg2+ are known to be involved in the recognition and strand exchange process (25).
For these reasons, we have first eliminated possible end effects by limiting the analysis to those predictions where the dsDNA interacts via its nine central base pairs (bases 8–16 and their complementary Crick bases). Within this selected set of geometries, a classification was made based on the interaction energy. In addition, we have concentrated the analysis on the predictions where the five central nucleotides (T5–T9) of the RecA-bound ssDNA contact at least one of these nine central bases of the incoming dsDNA, a contact being defined as a couple of ssDNA and dsDNA beads separated by less than 7 Å. Since the object of our study is the process of dsDNA/ssDNA sequence recognition within the scaffold of the nucleofilament, we are mainly interested in those geometries where the dsDNA is in close proximity of the ssDNA. The geometries of interest were therefore selected on the basis of the number of contacts NC between these two entities. Analysis was performed using the freely available object oriented library PTools developed in C++/Python [http://www.ibpc.fr/chantal/www/ptools (8)].
Interactive simulations
This part of the study was performed using the same coarse-grained representation as for systematic docking with ATTRACT, implemented in our interactive molecular simulation engine BioSpring. Flexibility is treated using elastic network models and the underlying simulation approach is fully described in the Supplementary Data 1. In the interactive simulations, the user can guide the docking process with a haptic device and thereby tune the direction and amplitude of forces applied during the run. User applied forces either act on single selected pseudo-atoms or on a multiple pseudo-atom selection. These forces are sent in real time to the simulation engine using the MDDriver library (12–15). Like for systematic docking, the system we studied was composed of five RecA monomers from the 3CMX crystal structure (5), this time with L2 loops present. Within these monomers, loops L2_0 to L2_4 were defined as flexible. These loops constitute the only interactively controlled protein region. Additionally, the DNA single strand of the nucleofilament was considered static, while the incoming dsDNA was flexible and guided by the interactive simulation. Control of the loop positions was obtained by successively pulling selected atoms, while the dsDNA position and orientation were more efficiently controlled by acting on a group of pseudo-atoms within the nine central Watson and Crick nucleobases (from 8 to 16 in one strand and from 32 to 40 in the complementary strand). Each single interactive simulation consisted in (i) moving L2 loops and simultaneously (ii) pulling the dsDNA toward the ssDNA, then (iii) allowing the relaxation of the system and finally (iv) saving the new state for further analysis. A total of 22 simulations were performed on the system, first to adjust the cutoff value used to build the elastic network, then to ensure a correct sampling. Details are given in Supplementary Data 1.
Coarse-grained to atomic model refinement
The passage from low to atomic resolution required the following steps. First, the atomic structures of both isolated L2 loops and dsDNA were, respectively, fitted on the corresponding low-resolution structures resulting from the interactive simulation, using the PTools library. For the loop structures, the fit was performed on the Cα positions while it was restricted to the phosphate atoms of nucleobases 8–16 and 32–40 for dsDNA. L2_3 was directly taken from the crystal structure. The C and N termini of each L2 loop L2_0 to L2_2 were then successfully connected back to the peptide main chain of the corresponding RecA monomers. For that purpose, we used the program Ligand (26,27) that allows the control of valence (for the main chain) and dihedral angles (main chain and side chains) using internal coordinates. The internal variables of the main chain where defined so that the Cα atoms of residues 202, 1202 and 2202, central to the L2 loops, were fixed in space. Skyhook types of restraints, where atom coordinates are constrained to predefined positions (here, directly taken from the crystal structure), were applied to the Cα, C and N atoms of residues 193 and 212, 1193 and 1212, 2193 and 2121, giving rise to an average deviation from the predefined coordinates of 0.45 Å.
The whole system was finally relaxed at atomic resolution using the sander module of the Amber9 program package (28). It involved 1000 steps steepest descent and 2000 steps conjugated gradient minimization followed by a short (4 ps) molecular dynamics simulation at 300 K and a second conjugated gradient energy minimization (2500 steps) to remove any residual steric overlap. Solvent effects were implicitly included using a distance-dependent dielectric function, with slope 4.
RESULTS
The coarse-grained DNA representation and force field used in this study have been designed and tested for assembling protein/DNA systems (6). Since we expect the ssDNA and dsDNA species to contact each other within the RecA filament, we performed a preliminary test to verify that these parameters can successfully reproduce DNA/DNA interactions Supplementary Data 2.
Docking dsDNA to RecA/ssDNA
This part of the study was performed in the absence of L2 loops. Our aim in using systematic simulations was to generate all possible positions and orientations of the encounter complex without any bias.
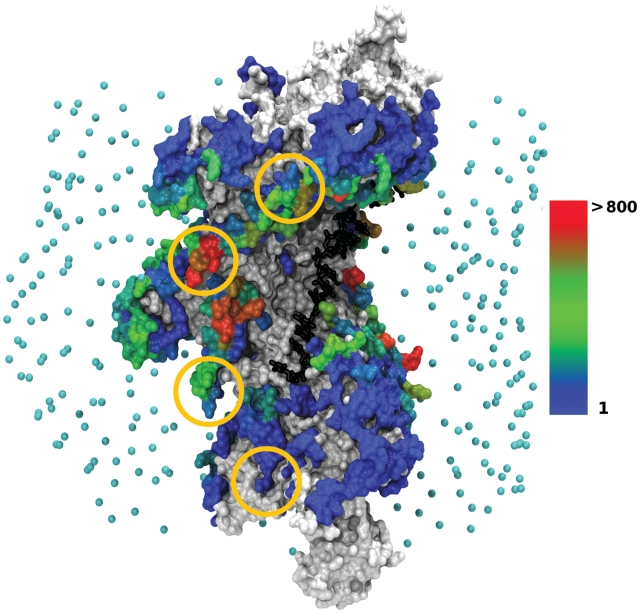
Whatever the DNA structure used for docking (either DSB or DSC), we verified that the filament surface was correctly sampled, except for the excluded extremities as explained in ‘Materials and Methods’ section. The most frequently occurring binding locations, which also correspond to the most favorable sites in terms of interaction energy, were found at the filament periphery, in sites repeatedly appearing in neighboring monomers (Figure 1). Interestingly, the DNA binding regions identified by the Kowalczykowski group based on cross-linking experiments (29) appear among the contacted regions (residues 233–243 of each monomer, marked by circles in Figure 1). Predictions where the dsDNA is fully inserted at the heart of the filament groove and contacts the ssDNA present interaction energies at least 2 RT above those involving the peripheric binding sites (Table 1). As discussed in the ‘Materials and Methods’ section, we did not expect these sites to show the best interaction energies because of the possible stabilizing role of missing loops L2. This role is investigated below using interactive simulations.
Figure 1.
Location of the dsDNA interaction sites on the filament/ssDNA surface, when dsDNA is the curved DSC structure. Analysis was performed on a set of 27 401 predicted geometries with interaction energies lower than −3.5 RT, where at least one contact is found between the central nine base pairs (8–16) and the filament. The protein filament is represented in surface mode constructed from the atomic representation. The ssDNA strand (black) is represented at atomic resolution in a stick representation. Points in cyan indicate the starting positions of the dsDNA center of mass in the docking simulations with ATTRACT. RecA amino acids are colored according to the number of times they are contacted by the dsDNA, cumulated over the set of predictions. White/gray patches correspond to contact-free amino acids. Amino acids contacted from 1 to 1 142 times are colored according to a color scale, from blue (once) to red (more than 800 times). Gold circles represent the location of experimentally identified DNA binding sites (29) (see text). Interaction sites corresponding to the docking of B-DNA can be found as Supplementary Data 3. The graphic representations in this figure and the following ones have been generated with VMD (30).
Table 1.
Results of docking simulations between dsDNA and RecA/ssDNA
| Prediction |
DSB |
DSC |
||
|---|---|---|---|---|
| Energy [RT] | NC | Energy [RT] | NC | |
| a | −7.3 | 0 | −12.3 | 0 |
| b | −5.3 | 1 | −9.0 | 1 |
| d | −1.2 | 3 | −6.9 | 5 |
Energy values (column 2 for DSB, 4 for DSC) and contact number NC (columns 3, 5) are reported for the docking predictions characterized by: (a) best energy; (b) best energy for the predictions with NC > 0; (d) highest NC number for a parallel orientation (see text).
B-DNA
Figure 2a presents a representative docking geometry between the 24 bp long DSB and the RecA/ssDNA filament. In this geometry, DSB contacts the ssDNA with a NC value of 3 (highest NC value for DSB, see Table 1). Typically, in the predictions with NC > 1, the ssDNA is found partly inserted in the major groove of DSB, the minor groove being probably too narrow to accommodate the ssDNA bases. The contacts mainly involve the bases of ssDNA and the phosphodiester backbone of DSB and extend over two or three bases. It can also be observed that the DSB base pairs are tilted with respect to the ssDNA bases by an angle of 30–45°. It is noteworthy that in the prediction represented in Figure 2a, as well as in Figure 2b for DSC, the 5′ branch of the dsDNA contacts the protein in one of the regions mentioned above, identified as possible DNA binding site by the group of Kowalczykowski (29) (Figure 1). This region may function as a hook point to keep the dsDNA close to the ssDNA during a period of time sufficiently long to allow for sequence probing.
Curved DNA
When DSC was used as ligand, the same docking protocol led to NC values up to 7, more than twice the maximum number of DSB/ssDNA contacts. In addition, the number of predictions with NC > 0 raised from 136 for DSB to 679 for DSC. The generated DSC positions that contact the ssDNA (NC > 0) can be divided in two groups presenting opposite orientations with respect to the RecA-bound ssDNA. The parallel orientation, where the two homologous strands are locally aligned in the same direction (Figure 2b), yields more favorable interaction energies between DSC and the filament (Table 1). Alternatively, the antiparallel orientation favors the number of DSC/ssDNA contacts over the interaction energy (−5.9 RT for NC = 7).
Figure 2b represents the geometry with parallel orientation which yields the most favorable interaction energy (−6.9 RT) and the highest NC value (NC = 5). The parallel orientation is required for strand exchange to occur, therefore we used this geometry in the following stages of the study. In that geometry as well as in all predicted geometries with NC > 1, the ssDNA contacts DSC in its minor groove. Contacts are limited to at most three consecutive base pairs interacting with three consecutive ssDNA bases.
Quite interestingly, the three bases (two bases from dsDNA and one from ssDNA) interacting at each of these three levels are oriented in an almost coplanar arrangement. This geometry strongly recalls, at the local level, the prestrand exchange configuration that we have proposed in a former work, based on the properties of stretched DNA (18,31). We will come back to this point in the ‘Discussion’ section.
Interactive investigation of L2 loop positions
To go further in the structural exploration of the early encounter complex, it was necessary to reintroduce the L2 loops lining the filament interior. As already explained, in the absence of incoming dsDNA, the loops fold upon the ssDNA, thus hindering its access to the searching dsDNA. This section describes the results of interactive simulations (14,15) starting from the structure presented in Figure 2b, where loops L2 taken from structure 3CMX have been reintroduced (‘Materials and Methods’ section). In these simulations, the L2 loops were flexible and the dsDNA was mobile. One haptic device was used to control the central nucleobases of the curved dsDNA, while a second haptic device was dedicated to independently control the motion of each L2 loop (see ‘Materials and Methods’ section and Supplementary Data 1; a short movie of the simulation can be seen at http://www.ibpc.fr/chantal/VR-RecA.m4v).
As expected, upon introduction of the L2 loops, we initially observed steric clashes between the loops and the dsDNA. These clashes could be eliminated by successively pulling loops L2_1 and L2_2 out while driving the dsDNA in close contact with the ssDNA. This yielded a system presenting a NC value of 25, five times higher than the initial value of 5 obtained from rigid body docking simulation. Upon relaxation, the displaced loop L2_1 got inserted in the major groove of the curved dsDNA, thus stabilizing its close approach. Interestingly, this structure of loop L2_1 associated with the best interacting dsDNA geometry occupies a spatial region close to that of the corresponding L2 loop in the crystal structure of MtRecA (Figure 3a). The 4.9 Å root mean square deviation (RMSD) value between the backbones of these two structures mostly reflects their difference in fold, the RMSD between both superposed structures being 3.0 Å. By comparison, the RMSD between the displaced L2_1 loop and its starting structure in the crystal form is 9.5 Å.
When we used DSB instead of DSC, it was again possible to drive the dsDNA close to the ssDNA by displacing two loops; however, we reached smaller NC values (up to 15). Note that for the highest NC values, the flexible dsDNA deformed to accommodate the single strand: its minor groove locally widened in the region contacting the ssDNA and small kinks (with an angle of ∼10°) appeared at each extremity of this region (data not shown). Such deformation resulted from user-applied external forces in the presence of steric constraints, but it may also reflect intrinsic mechanical properties of the double helix associated with minor groove widening (32).
Atomic details of the putative encounter complex
The passage from coarse-grained to atomic resolution required a phase of relaxation as described in the ‘Materials and Methods’ section, starting from the best structure obtained by interactive docking of DSC to the filament (previous section). The resulting structure, shown in Figure 3 and Supplementary Figure S4-1, depicts a densely packed complex where stabilizing interactions are found all along the incoming double helix. These interactions involve amino acids from four out of five consecutive RecA monomers and phosphodiester groups from both DNA strands. Interestingly, the central part of the dsDNA that is directly in contact with the ssDNA interacts almost exclusively through the backbone of the homologous strand, i.e. the strand which is homologous to the ssDNA, through a network of hydrogen bonding interactions and salt bridges with amino acids of loops L2_1 and L2_2 (Figure 3b and Supplementary Data 4). The aromatic side chain of Phe1203 (L2_1) also forms hydrophobic interactions in the major groove of the dsDNA and loop L2_0 presents an additional contact, this time with the complementary strand. Loops L1 (residues 2156–2165, 3156–3164), which already contribute to ssDNA binding, also offer functional groups for stabilization limited to the third and fourth monomers. The position of Met2164 in the minor groove of the double helix, close to a kink between base pairs T11.A38 and T12.A37, is worth noting since the same residues stabilize stacking interruptions observed every three base pairs in the RecA-bound duplex of the 3CMX crystal structure, by intercalating between unstacked base pairs of the complementary strand. In the perspective of a correspondence between kinks (for locally stretched DNA) and stacking gaps (for globally stretched DNA) (10), Met2164 is favorably positioned in the present structure for filling a gap in stacking that may form upon elongation of the ssDNA/dsDNA contact zone.
While RecA loops L2 and L1 constitute the main binding sites for the central part of the dsDNA, both dsDNA tails are anchored to the protein core in regions that partly recover the DNA binding locations identified by the Kowalczykowski group to [residues 233–243 (29)]. Contrarily to the central part, these interactions mainly involve the complementary strand. It is interesting to observe that the same amino acids Arg243, Lys245 and Gln254 from two consecutive monomers form hydrogen bonds with phosphodiester groups in the 3′ (first monomer) or the 5′-side (second monomer).
Comparison with the crystal structure of Chen et al. reveals a striking coincidence between the backbone of the dsDNA complementary strand in the model and the post-exchange DNA strand co-crystallized with the RecA/ssDNA nucleofilament in 3CMX (5) (respectively, red and black in Figure 3a). Both strands locally superpose, with identical orientations.
DISCUSSION
In the present study, we applied state-of-the-art modeling tools to investigate the possible arrangement of the actors of recognition, dsDNA, ssDNA, L2 loops and the protein scaffold, within the RecA nucleofilament. More precisely, we explored the possibility for the dsDNA to probe the ssDNA sequence while retaining its Watson–Crick connectivity. Due to the size of the system, the investigation was conducted at low resolution, followed by refinement at atomic resolution.
The search for homology is a crucial phase of HR, largely responsible for the efficiency of this fundamental process. In this phase, the whole genome of Escherichia coli is scanned by RecA nucleofilaments in a remarkably short period of time, on the order of minutes. Viovy and collaborators (33,34) showed that this short duration can be accounted for in the frame of a physical model, which includes polymer diffusion in a crowded environment and attributes different values to non-specific or specific nucleofilament/DNA interactions. In this model, initial recognition involves short segments that do not exceed 3 bp. These considerations imply a very short life time for unproductive encounter complexes, i.e. complexes where the aligned dsDNA and ssDNA fragments are not homologous. Indeed, kinetic studies have revealed high dissociation rate constants for the first stage of non-homologous complex formation, much greater than the association rate constant (35). For homologous DNA, Xiao et al. (36,37) measured a half-time in the order of seconds for the first encounter intermediate. A practical consequence is that it is experimentally very difficult to access the structure of the recognition intermediate. Thus, it is all the more important to gain insight into what happens during this crucial stage.
The known rapidity of homology search in the HR process together with the fact that ATP hydrolysis is not necessary for recognition and strand exchange (38) has also guided our choices all along the modeling process. As far as possible, we privileged solutions that avoid crossing high energy barriers. For example, we chose to investigate the association of the RecA-bound ssDNA to a dsDNA with intact pairing. In the same way, L2 loop displacements necessary for the dsDNA approach were modeled without altering the β-hairpin fold of these loops.
The results of our investigations call for a number of comments.
dsDNA structure
As expected, we have confirmed that the groove of the RecA nucleofilament is wide enough to accommodate a dsDNA. From a geometric point of view, the structure of the incoming DNA is not constrained by the groove topology: the DNA can be fully inserted into the groove both in its canonical form or in a curved form and it can approach the RecA-bound ssDNA in both forms during its search for sequence homology. In both cases, it can also anchor one extremity of its double helix into a region of the protein at the edge of the groove that was previously identified as a possible DNA binding site (29), while subsequent base pairs can search the inside of the filament groove.
Nevertheless, the characteristics of the nucleofilament/dsDNA interaction notably differ between the two tested forms. The overall interactions between the filament surface and the dsDNA clearly decrease when going from the curved to the straight forms (Supplementary Data 3). More specifically, the best interacting geometries where B-DNA approaches the ssDNA present few inter-DNA contacts and these contacts mostly involve the major groove side of the dsDNA and its phosphodiester backbone. This contrasts with several experimental proofs of a minor groove approach for the incoming DNA (39–42). Alternatively, when docking a curved dsDNA, the dsDNA and ssDNA oligonucleotides can enter in closer contact, with a more favorable interaction energy (Table 1), and the interaction is concentrated in the dsDNA minor groove. Similar results (data not shown) were obtained when docking a DNA structure built from the less distorted SRY-bound DNA structure (see Supplementary Data 3). Moreover, we observed that during flexible interactive docking simulations, the B-DNA structure could initiate a deformation toward the curved form, with a locally widened minor groove and the apparition of kinks at the junction between wide and narrow minor groove regions of the DNA.
While these observations support the scenario of a kinked incoming DNA structure within the association complex, the degree of curvature does not necessarily coincide with that of the TBP-bound TATA-box, which represents the most extreme deformation ever observed in crystal structures of DNA bound to architectural proteins. The fine structure of RecA-bound dsDNA certainly presents at least limited differences with the structure we chose as a model. The docking phase of our investigation should not suffer too much of this uncertainty, given that our low-resolution representation tolerates small to medium deformations of DNA [in previous tests, DNA structures with up to 20° bending deviation with respect to their form in the complex were correctly docked to their protein partners (6)].
The refinement phase showed the compatibility of the model DSC structure with the protein, and the possibility to form a whole network of favorable interactions, with the protein scaffold, simultaneously at both DNA extremities, and with the displaced L2 loops. However, we are conscious that the DNA structure needs to be further refined. This will require longer studies including dynamics at atomic resolution.
dsDNA/ssDNA interactions
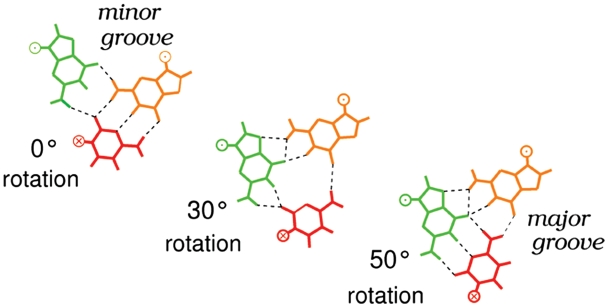
The geometric constraints favor a dsDNA orientation such that its interacting base pairs occupy a common plane with the ssDNA bases they contact. This arrangement noticeably resembles the pattern of minor groove interactions that was proposed in earlier work by Bertucat et al. (18,31). This work showed that such a geometry allows partial sequence recognition in the minor groove, limited to the GC containing bases, and that the bases are favorably positioned for pairing exchange. In this configuration, a simple 50° rotation of the complementary base of the duplex around its phosphodiester backbone is sufficient to induce the exchange of Watson–Crick partners (Figure 4). It is also worth recalling that in this former model, the stretched double helix structure that accommodates the ssDNA bases in its minor groove, as well as the double helix resulting from strand exchange, are remarkably similar to the recently solved crystal structure of the RecA-bound double helix (5,20). These remarks are supported by the close proximity between the backbone positions of the predicted complementary strand, in the DSC region in contact with ssDNA, and the corresponding strand in the crystal RecA-bound duplex (Figure 3a). All these elements converge to indicate that the docking prediction displayed in Figure 3 constitutes a plausible intermediate state both for sequence recognition and for pairing exchange.
Figure 4.
Simulation of the Watson–Crick pairing exchange within a G.C×G base triplet. Initially, the ssDNA guanine (green) interacts in the minor groove of the G.C base pair of the incoming dsDNA (orange and red, respectively, top view). The cytosine (red) is then rotated around its phosphodiester backbone while the rest of the system is allowed to relax. After 30° rotation, a ternary intermediate forms (middle) where all three bases interact with each other. After 50° rotation, the cytosine forms new Watson–Crick interactions with the ssDNA guanine (green) while the dsDNA guanine is displaced in the major groove of the newly formed heteroduplex (after the cover figure of Biophysical Journal, 1999, Vol.7, Issue 3).
L2 loops
Interactive experiments identified L2 loop geometries that not only allow, but also stabilize this type of geometry. We found that two consecutive L2 loops need to be displaced from their position in the crystal structure to allow close contact between the ssDNA bases and the minor groove of the dsDNA. Interestingly, loop geometries close to the position of L2 loops in MtRecA filaments (crystallized in the absence of DNA) appeared to allow stabilizing interactions in the major groove of the curved dsDNA. Since the corresponding spatial region is visited by L2 in the absence of ssDNA, we can reasonably expect that it constitutes an energetically favorable region that L2 can occupy during the dsDNA arrival.
What type of mechanism may drive the displacement of L2 loops is in itself an interesting question, as the interaction between these loops and the ssDNA in the crystal is sufficiently stable for their structure to be solved. A possible factor of destabilization may come from the perturbation of the potential energy in the filament groove during the approach of the dsDNA. Alternatively, it can be imagined that L2 loops undergo breathing movements that may momentarily clean the access to the ssDNA. Such a switching role has been previously proposed for loops L2 by the group of Camerini-Otero (43) in relation with the hydrolysis of ATP. Indeed, a scenario where continuous cycles of ATP hydrolysis would increase the frequency of L2 loop movements, thus accelerating the preliminary phase of dsDNA approach to the ssDNA, is appealing. However, no experimental data presently supports such hypothesis. To the contrary, the study by Gumbs and Shaner (44) comparing the kinetic steps of strand exchange in the presence of either ATP or its slowly hydrolyzed analog ATPγS did not detect any difference before the stage of strand exchange.
Implications for the HR mechanism
Finally, we will put our modeling results in the context of the discussions and hypotheses that have accompanied the progress in exploring the HR mechanism. Two scenarios have been envisioned for the recognition phase (45). In the first one, the dsDNA strands separate before encountering the ssDNA and the recognition simply consists in reading Watson–Crick functional groups. In the second scenario, the initial search contact between dsDNA and ssDNA occurs in one groove of the intact dsDNA. There are several arguments in favor of the first scenario. For example, the DNA stretching and unwinding deformations induced by its binding to RecA are known to favor base pair opening (46–48). However, former efforts to detect strand separation proved unsuccessful (49). In addition, a number of well-known examples exist where DNA can locally or globally stretch and unwind without loosing its base pairing (16,19,50,51), the last published example being the RecA-bound double helix solved by Chen et al. (5). More generally, the issue of DNA strand separation upon stretching is the object of a long-lasting controversy within the single molecule community (20). Gaub and collaborators (50) have recently proposed a cue to this problem, with an interpretation of their single molecule stretching experiments where two mechanisms compete. According to their results, DNA deformation under stretching load may lead to either DNA melting or DNA structural transition depending on the loading rate. At low rate, thermal fluctuations dominate and favor the melting process, while fast pulling leaves no time for thermal equilibration and results in a stretched S-DNA form with intact pairing. This interpretation makes the final form essentially dependent on the transition rate of distortion.
The second scenario involves a transient encounter complex where the dsDNA conserves its intact Watson–Crick pairing. Kinetic observations as well as theoretical modeling indicate that only partial recognition would occur within such a complex, via weak interactions between dsDNA and ssDNA bases (35,52). Complete recognition would require pairing exchange accompanied by direct Watson–Crick reading. This scenario is compatible with a quick dissociation of the encounter complex when no homology is detected (35). A mixed scenario is not excluded. It has been proposed by Radding and collaborators (53,54) as a result of studies on RecA or its human homolog Rad51, showing that A,T bases exchange their pairing as soon as they encounter. The kinetic studies of Singleton and collaborators (36,55) support the mixed scenario model.
The conditions of the present study, notably the use of a low-resolution DNA representation with rigidified base pairing during the early phase of systematic exploration, do not allow any conclusion on the melting scenario. Nevertheless, our results show that the second scenario is possible based on topological considerations. A paired dsDNA can transiently access the ssDNA bases for sequence probing via minor groove interactions, without previously loosing its base pairing. Our study shows that a curved form for the incoming dsDNA, apparented to the distorted structure induced by architectural proteins with locally widened minor groove, local unwinding and stacking interruptions (kinks), is perfectly adapted to such a successful approach. Its insertion in the nucleofilament groove is characterized by a minor groove approach, by initial alignment of three consecutive nucleotides for sequence probing and by a positioning of the dsDNA complementary strand favoring pairing exchange, with its backbone situated at the exact location of the X-ray complementary strand. This structure unexpectedly recalls a visionary scheme published two decades ago by Kubista et al. (56) based on their study of various RecA/DNA complexes using flow linear dichroism. It is perfectly consistent with the kinetic characteristics of the earliest recognition events established by Xiao et al. (36) in a remarkably comprehensive study. Notably, the authors noted that the activation enthalpy (of 33 kcal mol−1) associated with the first detectable intermediate of strand exchange is consistent with an initiation phase restricted to 3–4 bases, confirming the theoretical calculations of Dorfman et al. (34). Whether or not partial recognition occurs before pairing exchange, it is expected that the base pairs situated in the locally stretched region of the dsDNA will easily rotate toward the minor groove and open their pairing, thus initiating the strand exchange process (31,45). As noted by Radding and collaborators, this should be easier for A,T than for C,G sequences.
CONCLUSION
We have previously explored the possible contribution of the DNA mechanical properties to the recognition and strand exchange process (10). We have found that the transition to a stretched and unwound form, in phase with the RecA filament helicity, could by itself play an essential role in the process. The present study confirms that this putative role of the double helix geometry (limited here to a small portion) is compatible with its insertion in the RecA filament scaffold. By emphasizing the importance of L2 loop structural changes for both recognition and strand exchange, it also opens new perspectives regarding the possibility of an active role for RecA. The dynamics of the process now needs to be explored in a further study, using extensive molecular dynamics simulations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Agency for Research grants ANR-06-PCVI-0025 to O.D. and ANR-07-CIS7-003-01 (to C.A.); the Université franco-allemande (to A.S.); the Paris Diderot-Paris 7 University; the National Institute for Blood Transfusion (INTS); Institute for Health and Medical Care (Inserm) (to P.P.). Funding for open access charge: CNRS INTS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Takahashi, M. Dutreix, J.L. Viovy, R. Lavery and R. Egel for stimulating discussions, K. Bastard for early investigations on the RecA/ADP/DNA complex and M. Chavent for help with the figures.
REFERENCES
- 1.Camerini-Otero RD, Hsieh P. Homologous recombination proteins in prokaryotes and eukaryotes. Annu. Rev. Genet. 1995;29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- 2.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 3.Kuzminov A. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8461–8468. doi: 10.1073/pnas.151260698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson DG, Kowalczykowski SC. The recombination hot spot chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 6.Poulain P, Saladin A, Hartmann B, Prévost C. Insights on protein-DNA recognition by coarse grain modelling. J. Comput. Chem. 2008;29:2582–2592. doi: 10.1002/jcc.21014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zacharias M. Protein-protein docking with a reduced protein model accounting for side-chain flexibility. Protein Sci. 2003;12:1271–1282. doi: 10.1110/ps.0239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saladin A, Fiorucci S, Poulain P, Prévost C, Zacharias M. PTools: an opensource molecular docking library. BMC Struct. Biol. 2009;9:27–37. doi: 10.1186/1472-6807-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta S, Ganesh N, Chandra NR, Muniyappa K, Vijayan M. Structural studies on MtRecA-nucleotide complexes: insights into DNA and nucleotide binding and the structural signature of NTP recognition. Proteins. 2003;50:474–485. doi: 10.1002/prot.10315. [DOI] [PubMed] [Google Scholar]
- 10.Prévost C, Takahashi M. Geometry of the DNA strands within the RecA nucleofilament: role in homologous recombination. Q. Rev. Biophys. 2004;36:429–453. doi: 10.1017/s0033583504003956. [DOI] [PubMed] [Google Scholar]
- 11.Bastard K. PhD Thesis. Paris: Université Paris 7–Denis Diderot; 2005. Assemblage flexible de macromolcules: la théorie du champ moyen appliquée au remodelage des boucles protéiques. Assembling flexible macromolecules: The mean-field theory applied to the remodeling of protein loops. [Google Scholar]
- 12.Férey N, Delalande O, Grasseau G, Baaden M. A VR framework for interacting with molecular simulations. In: Kruiff E, editor. Proceedings of the 2008 ACM symposium on Virtual Reality Software and Technology. Bordeaux, France: ACM; 2008. pp. 91–94. [Google Scholar]
- 13.Férey N, Delalande O, Grasseau G, Baaden M. From interactive to immersive Molecular Dynamics. In: Faure F, Teschner M, editors. Proceedings of the International Workshop on Virtual Reality and Physical Simulation (VRIPHYS'08) Grenoble, France: Eurographics; 2008. pp. 89–96. [Google Scholar]
- 14.Delalande O, Férey N, Grasseau G, Baaden M. Complex molecular assemblies at hand via interactive simulations. J. Comput. Chem. 2009;30:2375–2387. doi: 10.1002/jcc.21235. [DOI] [PubMed] [Google Scholar]
- 15.Delalande O, Ferey N, Laurent B, Gueroult M, Hartmann B, Baaden M. Multi-resolution approach for interactively locating functionally linked ion binding sites by steering small molecules into electrostatic potential maps using a haptic device. Pac. Symp. Biocomput. 2010:205–215. doi: 10.1142/9789814295291_0023. [DOI] [PubMed] [Google Scholar]
- 16.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 17.Lebrun A, Shakked Z, Lavery R. Local DNA stretching mimics the distortion caused by the TATA box-binding protein. Proc. Natl Acad. Sci. USA. 1997;94:2993–2998. doi: 10.1073/pnas.94.7.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertucat G, Lavery R, Prévost C. A model for parallel triple helix formation by RecA: single-strand association with a homologous duplex via the minor groove. J. Biomol. Struct. Dyn. 1998;16:535–546. doi: 10.1080/07391102.1998.10508268. [DOI] [PubMed] [Google Scholar]
- 19.Lebrun A, Lavery R. Modeling DNA deformations induced by minor groove binding proteins. Biopolymers. 1999;49:341–353. doi: 10.1002/(SICI)1097-0282(19990415)49:5<341::AID-BIP1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 20.Prévost C, Takahashi M, Lavery R. Deforming DNA: from physics to biology. ChemPhysChem. 2009;10:1399–1404. doi: 10.1002/cphc.200900253. [DOI] [PubMed] [Google Scholar]
- 21.Lavery R. Junctions and bends in nucleic acids: a new theoretical modeling approach. In: Olson W, Sarma R, Sarma M, Sundaralingam M, editors. Structure and Expression, I : DNA Bending and Curvature. Schenectady, NY: Adenine Press; 1988. pp. 191–211. [Google Scholar]
- 22.Lavery R. Modelling the DNA Double Helix: Techniques and Results. In: Peyrard M, editor. Non-Linear Excitation in Biomolecules. Berlin and New York: Springer-Verlag; 1995. pp. 57–82. [Google Scholar]
- 23.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 24.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 25.Menetski JP, Bear DG, Kowalczykowski SC. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl Acad. Sci. USA. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bastard K, Thureau A, Lavery R, Prévost C. Docking macromolecules with flexible segments. J. Comput. Chem. 2003;24:1910–1920. doi: 10.1002/jcc.10329. [DOI] [PubMed] [Google Scholar]
- 27.Navizet I, Cailliez F, Lavery R. Probing protein mechanics: residue-level properties and their use in defining domains. Biophys. J. 2004;87:1426–1435. doi: 10.1529/biophysj.104.042085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Case D, Darden T, Cheatham T, Simmerling C, Wang J, Duke R, Luo R, Merz K, Pearlman D, Crowley M, et al. Technical report. San Francisco: University of California; 2006. Amber 9. [Google Scholar]
- 29.Rehrauer WM, Kowalczykowski SC. The DNA binding site(s) of the Escherichia coli RecA protein. J. Biol. Chem. 1996;271:11996–12002. doi: 10.1074/jbc.271.20.11996. [DOI] [PubMed] [Google Scholar]
- 30.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 31.Bertucat G, Lavery R, Prévost C. A molecular model for RecA-promoted strand exchange via parallel triple-stranded helices. Biophys. J. 1999;77:1562–1576. doi: 10.1016/S0006-3495(99)77004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zacharias M. Minor groove deformability of DNA: a molecular dynamics free energy simulation study. Biophys. J. 2006;91:882–891. doi: 10.1529/biophysj.106.083816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutreix M, Fulconis R, Viovy J-L. The search for homology: a paradigm for molecular interactions? Complexus. 2003;1:89–99. [Google Scholar]
- 34.Dorfman KD, Fulconis R, Dutreix M, Viovy J-L. Model of RecA-mediated homologous recognition. Phys. Rev. Lett. 2004;93:268102. doi: 10.1103/PhysRevLett.93.268102. [DOI] [PubMed] [Google Scholar]
- 35.Bazemore LR, Folta-Stogniew E, Takahashi M, Radding CM. RecA tests homology at both pairing and strand exchange. Proc. Natl Acad. Sci. USA. 1997;94:11863–11868. doi: 10.1073/pnas.94.22.11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao J, Lee AM, Singleton SF. Construction and evaluation of a kinetic scheme for RecA-mediated DNA strand exchange. Biopolymers. 2006;81:473–196. doi: 10.1002/bip.20446. [DOI] [PubMed] [Google Scholar]
- 37.Xiao J, Lee AM, Singleton SF. Direct evaluation of a kinetic model for RecA-mediated DNA-strand exchange: the importance of nucleic acid dynamics and entropy during homologous genetic recombination. ChemBioChem. 2006;7:1265–1278. doi: 10.1002/cbic.200600038. [DOI] [PubMed] [Google Scholar]
- 38.Rosselli W, Stasiak A. Energetics of RecA-mediated recombination reactions: without ATP hydrolysis RecA can mediate polar strand exchange but is unable to recycle. J. Mol. Biol. 1990;216:333–350. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- 39.Baliga R, Singleton JW, Dervan PB. RecA.oligonucleotide filaments bind in the minor groove of double-stranded DNA. Proc. Natl Acad. Sci. USA. 1995;92:10393–10397. doi: 10.1073/pnas.92.22.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Podyminogin MA, Meyer RB, Gamper HB. RecA-catalyzed, sequence-specific alkylation of DNA by cross-linking oligo-nucleotides. Effects of length and nonhomologous base substitutions. Biochemistry. 1996;35:7267–7274. doi: 10.1021/bi9529465. [DOI] [PubMed] [Google Scholar]
- 41.Zhou X, Adzuma K. DNA strand exchange mediated by the Escherichia coli RecA protein initiates in the minor groove of double-stranded DNA. Biochemistry. 1997;36:4650–4661. doi: 10.1021/bi9630063. [DOI] [PubMed] [Google Scholar]
- 42.Singleton SF, Xiao J. The stretched DNA geometry of recombination and repair nucleoprotein filaments. Biopolymers. 2001;61:145–158. doi: 10.1002/bip.10145. [DOI] [PubMed] [Google Scholar]
- 43.Voloshin ON, Wang L, Camerini-Otero RD. The homologous pairing domain of RecA also mediates the allosteric regulation of DNA binding and ATP hydrolysis: a remarkable concentration of functional residues. J. Mol. Biol. 2000;303:709–720. doi: 10.1006/jmbi.2000.4163. [DOI] [PubMed] [Google Scholar]
- 44.Gumbs OH, Shaner SL. Three mechanistic steps detected by FRET after presynaptic filament formation in homologous recombination. ATP hydrolysis required for release of oligonucleotide heteroduplex product from RecA. Biochemistry. 1998;37:11692–11706. doi: 10.1021/bi980646s. [DOI] [PubMed] [Google Scholar]
- 45.Howard-Flanders P, West SC, Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984;309:215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 46.Bernet J, Zakrzewska K, Lavery R. Modelling base pair opening: the role of helical twist. J. Mol. Struct. THEOCHEM. 1997;398-399:473–482. [Google Scholar]
- 47.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix 1. Thermodynamic analysis. Biophys. J. 2001;80:882–893. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris SA, Sands ZA, Laughton CA. Molecular dynamics simulations of duplex stretching reveal the importance of entropy in determining the biomechanical properties of DNA. Biophys. J. 2005;88:1684–1691. doi: 10.1529/biophysj.104.046912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsieh P, Camerini-Otero RD. Formation of joint DNA molecules by two eukaryotic strand exchange proteins does not require melting of a DNA duplex. J. Biol. Chem. 1989;264:5089–5097. [PubMed] [Google Scholar]
- 50.Albrecht CH, Neuert G, Lugmaier RA, Gaub HE. Molecular force balance measurements reveal that double-stranded DNA unbinds under force in rate-dependent pathways. Biophys. J. 2008;94:4766–4774. doi: 10.1529/biophysj.107.125427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danilowicz C, Limouse C, Hatch K, Conover A, Coljee VW, Kleckner N, Prentiss M. The structure of DNA overstretched from the 5′5′ ends differs from the structure of DNA overstretched from the 3′3′ ends. Proc. Natl Acad. Sci. USA. 2009;106:13196–13201. doi: 10.1073/pnas.0904729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertucat G, Lavery R, Prévost C. A mechanism for RecA-promoted homology recognition and strand exchange between single-stranded DNA and duplex DNA via triple-helical intermediates. In: Sarma R, Sarma M, editors. Proceedings of the XIth Conversation in the Discipline Biomolecular Stereodynamics. Vol. 1. Albany, NY, USA: Adenine Press; 2000. pp. 147–152. [DOI] [PubMed] [Google Scholar]
- 53.Folta-Stogniew E, O'Malley S, Gupta R, Anderson KS, Radding CM. Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein. Mol. Cell. 2004;15:965–975. doi: 10.1016/j.molcel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 54.Gupta RC, Folta-Stogniew E, O'Malley S, Takahashi M, Radding CM. Rapid exchange of A:T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol. Cell. 1999;4:705–714. doi: 10.1016/s1097-2765(00)80381-0. [DOI] [PubMed] [Google Scholar]
- 55.Lee AM, Xiao J, Singleton SF. Origins of sequence selectivity in homologous genetic recombination: insights from rapid kinetic probing of RecA-mediated DNA strand exchange. J. Mol. Biol. 2006;360:343–359. doi: 10.1016/j.jmb.2006.04.065. [DOI] [PubMed] [Google Scholar]
- 56.Kubista M, Takahashi M, Nordén B. Stoichiometry, base orientation, and nuclease accessibility of RecA.DNA complexes seen by polarized light in flow-oriented solution. Implications for the mechanism of genetic recombination. J. Biol. Chem. 1990;265:18891–18897. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.