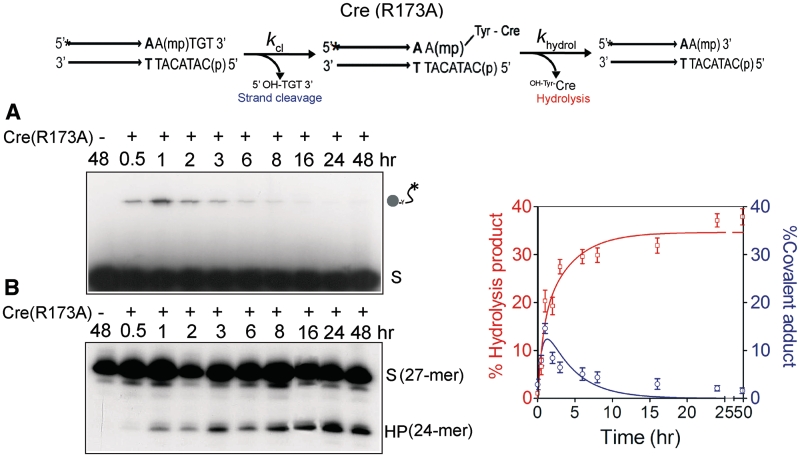
Figure 2.
Strand cleavage and type I endonuclease activities of Cre(R173A) on an MeP-half-site substrate. In the schematic representation of the half-site substrate, the asterisk indicates 32P-label at the 5′-end, and ‘mp’ the scissile methylphosphonate. The parallel arrows denote the Cre binding element. Short stretches of additional nucleotides extending from the binding element to its left on both strands are not shown. The type I endonuclease activity proceeds via formation of the MeP–tyrosyl intermediate, followed by its hydrolysis. Reactions were split into halves to analyze the covalent Cre–DNA adduct (indicated by the circular knob linked to the labeled DNA chain) by SDS–PAGE (A) and the potential hydrolysis product(s) by denaturing urea–PAGE (B). The end-labeled half-site (A) or its labeled strand (B) is denoted by ‘S’, and the hydrolysis product by ‘HP’. The labeled strand in the MeP-half-site is 27-nt long, and HP is a 24-mer. In the control reaction, the MeP-half-site was incubated in the recombination buffer for 48 h in the absence of Cre(R173A). The plots represent mean values from three separate experiments with Cre(R173A). The global fit to the kinetic data was performed using KinTek Global Kinetic Explorer, as explained in ‘Materials and Methods’ section. For a given starting concentration of the MeP-half-site, each rate constant and the output scaling factor were adjusted to obtain the optimal global fit.