Table 2.
Activities of Cre mutants containing alanine replacements at Arg-173 or Arg-292 or both positions deduced from this study are summarized
| Cre mutant | Activities on MeP DNA substrates |
||||
|---|---|---|---|---|---|
| Strand cleavage | Type I endonuclease (hydrolysis of MeP–tyrosyl intermediate) | Type II endonuclease (direct hydrolysis of MeP bond in DNA) | Strand joining | Recombination | |
| Cre(R173A) | Active (kcl = 4.4 × 10−4s−1) | Active (khydrol = 2.7 × 10−4s−1) | Inactive | Active (kjoin = 8.6 × 10−6s−1) | Active |
| Cre(R292A) | Active (kcl = 3.2 × 10−4s−1) | Active (kcl = 8.8 × 10−4s−1) | Inactive | Active (kjoin = 2.8 × 10−5s−1) | Active |
| Cre(R173A, R292A) | Active 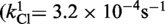 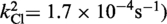
|
Active 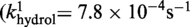 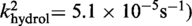
|
Inactive | Inactive | Inactive |
The kinetic data for the strand cleavage and endonuclease activities of Cre(R173A, R292A) yielded better global fit by assuming two contributing first-order rate constants for each activity. The distinct reactivities of the RP and SP forms of the MeP–substrate (provided as an achiral mixture of the two forms) could potentially account for the binary rate constants.
