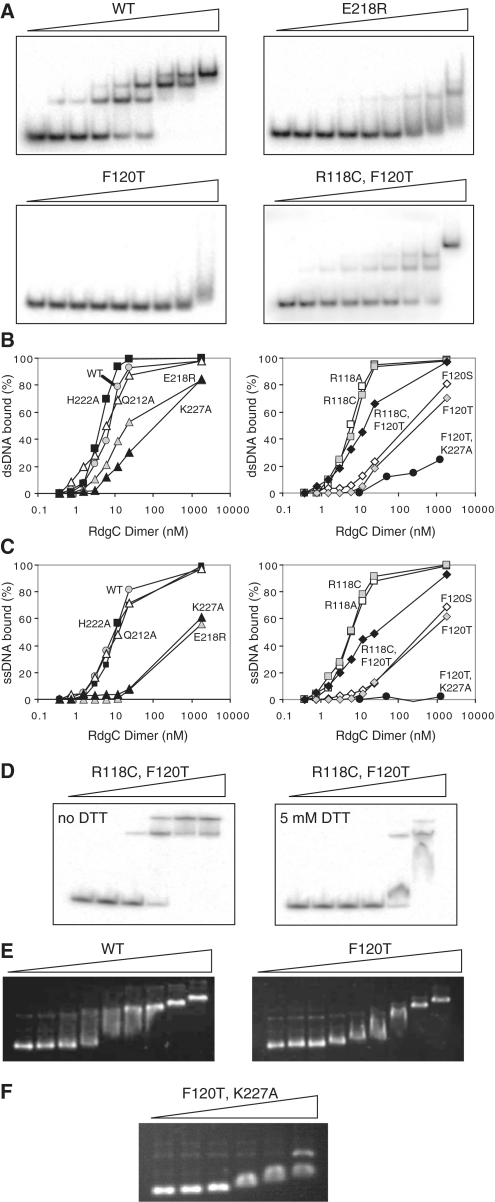
Figure 4.
Oligonucleotide binding by RdgC proteins. (A) Representative band-shift assays with dsDNA. Reactions used the RdgC proteins indicated at 0, 0.375, 0.75, 1.5, 3, 6, 12, 24 and 1850 nM (of dimer) and 32P-labelled RGL13/17 DNA at 0.2 nM. Assays with R118A, R118C, Q212A and H222A gave the same band pattern as the WT protein. Assays with F120S and K227A gave the same band pattern as F120T. (B) Quantification of dsDNA binding. RGL13/17 DNA was used at 0.2 nM. Data are means of at least two band-shift experiments. (C) Quantification of ssDNA binding. 32P-labelled RGL13 DNA was used at 0.2 nM. Data are means of at least two band-shift experiments. (D) Effect of DTT on duplex binding by RdgC R118C F120T. Reactions used mutant RdgC protein at 0, 1, 5, 25, 125 and 625 nM (of dimer) and 32P-labelled RGL13/17 DNA at 0.2 nM. (E) Plasmid binding by RdgC proteins. Reactions used the RdgC proteins indicated at 0, 34, 68, 136, 272, 544, 1088, 2176 and 23 400 nM (of dimer) and pGEM-7 Zf(+) circular dsDNA at 2.2 nM plasmid (6480 nM nucleotide pairs). (F) Plasmid binding by the F120T K227A double mutant. Reactions used mutant RdgC at 0, 1000, 2000, 5000, 10 000 and 20 000 nM (of dimer) and pGEM-7 Zf(+) circular dsDNA at 2.2 nM plasmid (6480 nM nucleotide pairs).