Figure 1.
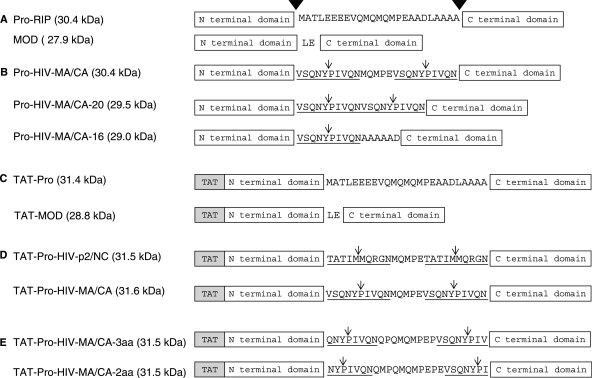
Schematic diagram of the maize RIP and recombinant variants. (A) The artificial construct maize RIP precursor Pro-RIP (Pro-RIP) contains the 25 aa internal inactivation region but without the 16 aa N-terminal and 14 aa C-terminal sequences of the inactive maize RIP precursor. The amino acid sequence of the internal inactivation region is shown. Cleavage sites leading to the generation of the active two-chain maize RIP are indicated by inverted filled triangles. MOD is another artificial construct with the two polypeptides fused by an Leu-Glu bipeptide. (B) The internal inactivation region was replaced by HIV-1 protease recognition site. Pro-HIV-MA/CA: the first and last 10 aa of the internal inactivation region were replaced by the 10 aa MA/CA site. Pro-HIV-MA/CA-20: two 10 aa MA/CA sites were introduced to the internal inactivation region. Pro-HIV-MA/CA-16: the whole internal inactivation region was replaced by one copy of MA/CA site followed by a hexapeptide of AAAAAD. (C) TAT-Pro and TAT-MOD: A Tat sequence was fused to the N-termini of Pro-RIP and MOD, respectively. (D) TAT-Pro-HIV-p2/NC and TAT-Pro-HIV-MA/CA: TAT-Pro with the first and last 10 aa replaced by the p2/NC site (TATIM/MQRGN) and the MA/CA site (VSQNY/PIVQN), respectively. (E) TAT-Pro-HIV-MA/CA-2aa and TAT-Pro-HIV-MA/CA-3aa: the recognition sequence of the MA/CA site was shortened to reduce the number of aa left to 2 and 3, respectively, after cleavage. The underlined sequences are the recognition sites of HIV-1 protease and ↓ indicates the HIV-1 protease cleavage site.