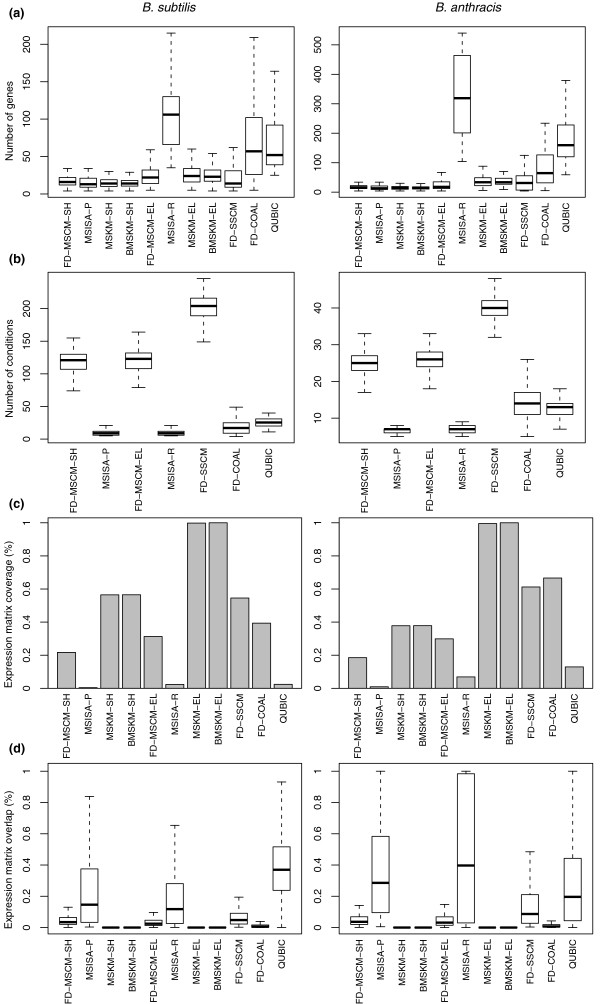
Figure 3.
Comparison of the size, coverage and overlap for single and multi-species methods for the B. subtilis-B. anthracis pairing (full data results only, where applicable). For brevity, we present here only the results from full data methods (FD) from the B. subtilis-B. anthracis pairing (results for the other pairings and EO methods can be found in Additional file 1). (a) The distribution of the number of genes in the (bi)clusters from the different methods. There is a consistent increase in the median size between the shared and elaboration steps (this is most extreme in the case of the MSISA method). For both organisms, Coalesce (COAL) and QUBIC produced the next largest biclusters, in terms of the number of genes. (b) The distribution of the number of conditions in the biclusters from the different biclustering methods only. We do not show this for the MSKM and BMSKM results as these methods use all conditions. For both organisms, the MS/SS cMonkey methods produced the biclusters with the most conditions. The MSISA method produced the biclusters with the least number of conditions. (c) The coverage of the total expression data matrix by the (bi)clusters from the different methods is displayed. The elaborated results of the MSKM and BMSKM methods achieve perfect coverage, by definition. The MSISA and QUBIC biclusters had the smallest coverage of any of the methods, while the Coalesce biclusters achieved coverages comparable with the SSCM biclusters. (d) The distribution of all pairwise, non-zero overlaps between the (bi)clusters from the different methods; overlap in terms of the overlap of expression matrix elements, rather than genes. By definition, the MSKM and BMSKM clusters have no overlap, while the MSISA and QUBIC biclusters had the greatest. Of the biclustering methods, Coalesce had the least overlap. Coalesce identifies more distinct biclusters with greater numbers of genes, but fewer conditions; and the SS/MS cMonkey methods identify biclusters that are slightly more overlapped than does Coalesce, with fewer genes, but covering more conditions.