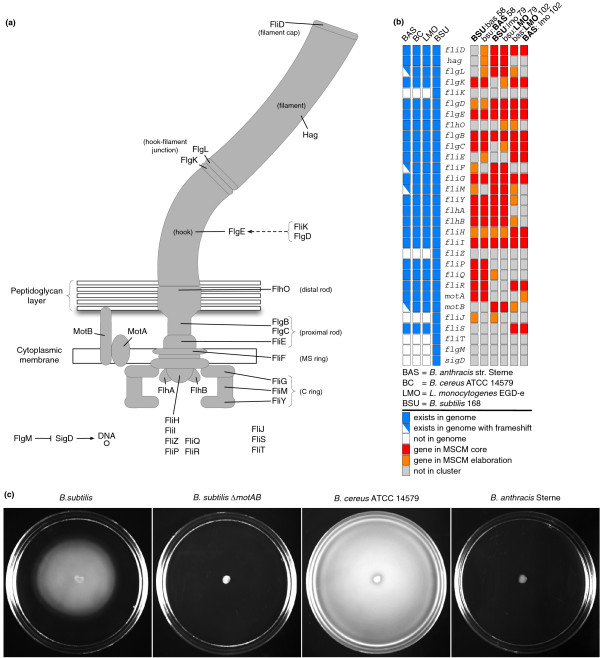
Figure 6.
Conserved motility modules active in all three organisms and motility assays. (a) We show a schematic of the flagellar apparatus for B. subtilis showing the location of 26 flagellar proteins, two motor proteins (MotA and MotB) and two transcriptional regulators (FlgM and SigD) (using gene names from B. subtilis). (b) The left panel shows the presence (blue)/absence (white) of the corresponding genes in the genomes of B. anthracis Sterne (BAS), B. cereus ATCC 14579 (BC), L. monocytogenes EGD-e (LMO) and B. subtilis 168 (BSU). In B. anthracis Sterne, motB, fliM, fliF, and flgL are represented by two colors indicating a gene coding for a truncated protein due to a frameshift mutation that introduces a premature stop codon. The right panel shows the gene presence in the main flagellar bicluster resulting from each of the three pairwise multi-species biclusterings. Indicated are genes of the flagellar apparatus - included in the bicluster core (red), in the elaboration of the bicluster (orange), and not included in the bicluster (gray). B. subtilis and L. monocytogenes are both known to be flagellated and motile. B. anthracis Sterne is non-motile, but our results indicate a bicluster enriched for genes involved in flagellar biosynthesis. (c) Swimming motility was assayed on 0.3% agar plates for B. cereus ATCC 14579, B. anthracis Sterne, B. subtilis PY79, and B. subtilis PY79 ΔmotAB::tet (strain DS219). B. cereus and B. subtilis are motile [51,68]. A deletion of motAB in B. subtilis impairs motility [69,91]. The assay shows that B. anthracis Sterne is not motile under the conditions tested.